Abstract
Timely detection and treatment of microaneurysms (MA) is a critical step to prevent the development of vision-threatening eye diseases such as diabetic retinopathy. However, detecting MAs in fundus images is a highly challenging task due to the large variation of imaging conditions. In this paper, we focus on developing an interleaved deep mining technique to cope intelligently with the unbalanced MA detection problem. Specifically, we present a clinical report guided multi-sieving convolutional neural network (MS-CNN) which leverages a small amount of supervised information in clinical reports to identify the potential MA regions via a text-to-image mapping in the feature space. These potential MA regions are then interleaved with the fundus image information for multi-sieving deep mining in a highly unbalanced classification problem. Critically, the clinical reports are employed to bridge the semantic gap between low-level image features and high-level diagnostic information. Extensive evaluations show our framework achieves 99.7% precision and 87.8% recall, comparing favorably with the state-of-the-art algorithms. Integration of expert domain knowledge and image information demonstrates the feasibility to reduce the training difficulty of the classifiers under extremely unbalanced data distribution.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Diabetic retinopathy (DR) is the leading cause of blindness globally. Among an estimated 285 million people with diabetes mellitus worldwide, nearly one-third have signs of DR [1]. Fortunately, the risk of vision loss caused by DR can be notably reduced by early detection and timely treatment [2]. Micro-aneurysm (MA), the earliest clinical sign of DR, is defined as a tiny aneurysm occurring secondary to the capillary wall and appear as small red dots in the superficial retina layers. MA counts is an important measure of progression of retinopathy in the early stage and may serve as a surrogate endpoint for severe change in some clinical trials [3]. However, manual segmentation and counting of MA is time-consuming, subjective, error-prone and infeasible for large-scale fundus image analysis and diagnose. Therefore automatic detection and counting of MA is a core component of any computer-aided retinopathy diagnosis system.
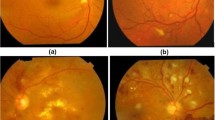
However, several factors, including the variation in image lighting, the variability of image clarity, the occurrence of another red lesion, extremely low contrast and highly variable image background texture, made the segmentation of MA difficult for an automated system. Figure 1 shows some examples of fundus images containing challenging MAs for detection.
Difficult cases in MA detection. (a) A normal and obvious MA. (b) Blood vessel joints similar to MA (blue arrow). (c) Light and texture varies. (d) Hemorrhage (green arrow) may cause false positive detection (e) Blurred fundus image, which makes of MA (white arrow) detection more difficult. (f) Reflection noise (yellow arrow). (Color figure online)
To address the above challenges, we proposed a multi-modal framework utilizing both expert knowledge from text reports and color fundus images. Different from previous methods, our proposed framework is able to (1) integrate non-image information from experts through clinical reports; and (2) accommodate the highly unbalanced classification problem in medical data.
2 Methodology
Our framework consists of two phases. In Phase 1, a statistical image-text mapping model is generated using the algorithm described in Sect. 2.1. The model maps visual features to different lesion types in the retina. The learned image-text mapping model will generate a lesion estimation map, where features of superpixels in the fundus images can be easily decoded to types and confidence of lesions via the lesion estimation map. In Phase 2, we propose multi-sieving convolutional neural networks (MS-CNNs), as described in Sect. 2.2, to perform pixel-level binary lesion classification in fundus images. The challenge is the highly unbalanced data where the number of pixels with lesions can be only one-thousandth of that of healthy pixels. The color information from the raw fundus images is coupled with the lesion estimation map from Phase 1, and fed into MS-CNN for MA detection.
2.1 Learning Image-Text Mapping Model
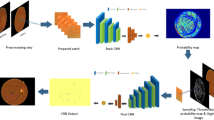
In this section, we adapt the image-text mapping model introduced in [4] to extract expert knowledge from clinical reports. The input into the model is the fundus images with their corresponding clinical text reports, and the output of this model is a probabilistic map of the lesion types in the fundus image. Our proposed image-text mapping model consists of five stages, as shown in Fig. 2: (1) image preprocessing and feature extraction; (2) text information extraction from clinical reports; (3) random feature space partitioning; (4) lesion distribution model; (5) mapping features to lesion types.
The illustration of learning the image-text mapping model. The top panel illustrates the training procedure of the image-text mapping model. A text-image mapping model is learned through the feature space partitions’ lesion label distribution (middle panel is inspired by Hofmanninger and Langs’ work [4]). At test time, images are segmented into superpixels which are also mapped to the feature space for lesion label prediction (bottom panel).
First, we resize all image to the same size and apply histogram balance to images to eliminate the large variation in imaging conditions. To improve the computational efficiency, the fundus images are over-segmented into superpixels using Simple Linear Iterative Clustering (SLIC) [5]. Features are extracted from superpixels using a pre-trained AlexNet model provided in Caffe Model Zoo, where we use the fully connected neural network layer of AlexNet [6] which has a high representation power of image features. We use a rectangle patch at the center of superpixels as the input into CNN because the shape of the superpixels are not regular.
Then, we extract the lesion information from the clinical text reports written in natural languages. Based on our observation, clinical text reports always contain the names of lesions appearing in the corresponding fundus images and in general lesions are only mentioned when they exist. So we represent the keywords of lesion names from clinical text reports as binary arrays, with examples shown in Table 1.
Next, given the image visual features and the corresponding lesion types in fundus images from the previous steps, we partition the visual feature space by assigning probability weights of each lesion type to each feature space partition. Because the location information of the lesions is missing from the text reports, a semantic mapping from visual features [4] is utilized to fill in this gap. To this end, the feature space is first randomly and uniformly partitioned using random fern [7] which generates an index value for each subspace. A fern is a sequence of binary decision functions \(\mathcal {F}\). These functions map the feature vectors of superpixels to an integer index of the partitioned space. Suppose there are L binary functions in \(\mathcal {F}\), as each binary function \(\mathcal {L}_i\,(i=1,\ldots ,L)\) produces a bit in \(\mathcal {F}\), the resultant binary code represents values ranged from 1 to \(2^{L}\). The mapping between the superpixels and lesion types is obtained by assigning the lesion types in the clinical reports to all superpixels in the corresponding fundus image.
Inspired by term frequency-inverse document frequency (tf-idf) model in natural language processing [8] and Hofmanninger and Langs’ method [4], we developed a model called “partition frequency-inverse lesion frequency” (pf-ilf model) to identify the best feature partitions to represent each lesion type. Here lesion types are treated as documents and feature space partitions as terms. We use Laplacian smoothing to avoid zero partition frequency for some lesion types. Inverse lesion frequency ilf for partition p is defined as the total number of lesion types divided by the number of lesions that fall in the partition p. Then we can define the score pf-ilf of a partition p for a lesion type l as \(\text {pd-ilf}(p,l) = \frac{1+f_{p,l}}{L+\max _{k} f_{k,l}}\times log_2\frac{L}{n_p}\), where \(f_{p,l}\) is the number of superpixels with the lesion label l that fall in the feature space partitions p. L is the total number of lesion types, \(n_p\) is the number of lesions that fall into partition p. With the proposed lesion distribution model using pf-ilf score, we can identify the most representative feature space partitions p for a specific lesion type l, by ranking the pf-ilf scores of all partitions for the lesion type l. The middle panel of Fig. 2 visualizes the random subspace partitioning of the visual feature space and the mapping between feature space partitions and lesion types.
Finally, we predict the lesion types in each superpixel using the image-text mapping model, as illustrated in the middle and bottom panels of Fig. 2. For each lesion type l, we pick the top k partitions with highest pf-ilf defined as \(P_l(k)\). Suppose a superpixel s is mapped to a set of partitions \(Q_s\) in the feature space. We define a final score of a superpixel s and a lesion type l as \(S(s,l)=|P_l(k)\cap Q_s|\), indicating the size of the intersection set between \(P_l(k)\) and Q(s). Finally the superpixel s is labeled with the lesion type l with the highest score S(s, l).
2.2 Multi-Sieving Convolutional Neural Network for MA Detection
In spite of its efficacy in large-scale image segmentation and object detection [6], CNN still faces limitations when dealing with unbalanced data, such as detecting the MAs in fundus images. Firstly, it favors balanced datasets, while the ratio of positive examples (MA) to negative examples (non-MA) can be as low as 1:1000. Second, multiple misleading visual traits such as blood vessels can lead to erroneous classification using only visual features of the fundus image. In other words, non-image information would provide critical meta-data to guide the classification model by integrating additional cues such as expert knowledge from the clinical reports. The right image in the bottom panel of Fig. 2 visualizes the clinical report guided segmentation from the image-text mapping model.
To address the unbalanced data challenge, we first propose Multi-Sieving Convolutional Neural Networks (MS-CNNs) to address the unbalanced dataset issue. MS-CNNs are a cascaded CNN model with the false positives from the previous network fed into the next network as negative examples. Suppose all positive samples are in set P and negative samples are in set N. For the first phase, we select all samples in P and randomly select an equal number of samples in N as initial training samples \((P^{(0)}, N^{(0})\). Then for the \(n^{th}\) phase, we first perform classification using network trained in previous phase on all samples in \(P^{(n-1)}\) and \(N^{(n-1)}\). This will generate many false positive predictions which will be collected in a set \(FP^{(n)}\). We select all positive samples in P again, but now randomly select an equal number of negative samples from \(FP^{(n)}\), which are “hard” or challenging samples for the previous classifier.
3 Experimental Results
In this section, extensive experiments are carried out to evaluate the clinical report guided multi-sieving CNN model for MA detection. We collect a dataset including fundus images and clinical reports from a local hospital. This dataset is used for training and testing the image-text mapping model. We also use the standard diabetic retinopathy (DIARETDB1 [9]) database to test our clinical report guided MS-CNN model.
(a) Illustration of expert knowledge learning result. The top row is the original fundus images with different kinds of lesions. The bottom row is the corresponding output of image-text mapping model. The original output images are gray level images where different gray levels represent different lesion type. We transformed the original output images to pseudo-color images for visualization purpose. (b) Illustration of segmentation results of the first and second blocks of MS-CNN. White, red, and green dots represent false positive, false negative, and true positive predictions respectively. (Color figure online)
The dataset collected from the local hospital contains 646 images. All images have a resolution of 3504 \(\times \) 2336. 433 of them are without obvious DR and the rest images contain different lesion types. The DIARETDB1 dataset contains 89 non-dilated fundus images with different types of lesions. We evaluate the efficacy of our proposed framework in terms of classification precision and recall.
All of our experiments were performed using the CNN with the same configuration as AlexNet [6]. We train and test the image-text mapping model using the dataset collected from the local hospital. 80% of images are randomly selected as the training set and the rest for testing. Two annotators annotated disjoint subsets of the local dataset based on the fluorescence fundus angiography. Because text reports are not available in DIARETDB1, we use the same image-text mapping model trained using the local data to perform tests on the DIARETDB1 dataset. Following the experiment setup in [9], 28 images in the dataset are pre-selected as the training data and the rest images are used as the testing data. To show that the extra channel helps the MA detection, we trained and tested two CNNs with the same architecture, but one with the extra expert knowledge-guided channel, one with the extra channel filled with zeros to avoid CNN structural change.
To compare our method with existing approaches, we implemented, turned and tested two state-of-the-art methods described in [10, 11]. We also compared with three methods published recently [12,13,14]. Our method achieved the highest score in both recall and precision compared to all other methods, with comparable accuracy to [13]. We also observe from Table 2 that without the clinical report guided information, the proposed MS-CNN method already outperforms all state-of-the-art methods in terms of recall. Furthermore, with the clinical report guided information, our proposed method achieved the highest precision compared with the best of the five methods [13]. We also observe that there is a substantial increase of recall from 84.2% to 87.8% with a significant 5.7% improvement when image-text mapping channel is added. This is critical to medical image analysis, as false negatives can be detrimental for the disease diagnosis. This indicates that some MAs are too vague to be distinguished from background. Our image-text mapping model is able to find the right properties of MA and thus provide necessary information to CNN. We also expected a significant increase in precision, but only a slight increase from 98.8% to 99.7% is observed. We believe that the multi-sieving scheme eliminated most of the false positive predictions. More blocks in MS-CNN have also been experimented, but the improvement in performance is negligible while the computational cost increases linearly with the number of blocks. So we keep 2 blocks for the MS-CNN framework (Fig. 3).
To demonstrate that multi-sieving scheme is effective, we extracted the results from the first and the second block of MS-CNN. As expected, the recall increases sharply from 17.9% using only 1 block to 87.7% using 2 blocks. But we also noticed that precision decreases slightly from 100% to 99.7%. We have to tradeoff between precision and recall. Since the main purpose of MS-CNN is to increase recall, a slight decrease of precision is acceptable and overall performance is improved.
4 Conclusions
The paper presents a novel clinical report guided framework for automatic microaneurysm detection from fundus images. We first extract expert knowledge from clinical text reports and map visual features to semantic profiles. Integration of keywords information from text reports and features from fundus images help to boost the detection accuracy with a promising performance in terms of precision and recall. The proposed framework performs favorably by overcoming MA detection challenges including unbalanced dataset and varying imaging conditions faced by the existing approaches. This is mainly achieved by multimodality information integration from clinical reports and visual features, and a multi-sieving classification strategy. The framework proposed in this paper is a generic approach that can be easily extended to detection of multiple kinds of lesions in fundus images and other medical imaging modalities such as octical coherence tomography (OCT) and angiography.
Change history
15 February 2019
In the originally published version important references were omitted.
References
Lee, R., Wong, T.Y., Sabanayagam, C.: Epidemiology of diabetic retinopathy, diabetic macular edema and related vision loss. Eye Vis. 2(1), 1 (2015)
UK Prospective Diabetes Study Group. Tight blood pressure control and risk of macrovascular and microvascular complications in type 2 diabetes: Ukpds 38. BMJ: British Medical Journal, pp. 703–713 (1998)
Klein, R., Meuer, S.M., Moss, S.E., Klein, B.E.K.: Retinal microaneurysm counts and 10-year progression of diabetic retinopathy. Arch. Ophthalmol. 113(11), 1386–1391 (1995)
Hofmanninger, J., Langs, G.: Mapping visual features to semantic profiles for retrieval in medical imaging. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 457–465 (2015)
Achanta, R., Shaji, A., Smith, K., Lucchi, A., Fua, P., Susstrunk, S.: SLIC Superpixels. EPFL Technical report 149300, p. 15, June 2010
Krizhevsky, A., Sulskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. In: Advances in Neural Information and Processing Systems (NIPS), pp. 1–9 (2012)
Ozuysal, M., Calonder, M., Lepetit, V., Fua, P.: Fast keypoint recognition using random ferns. IEEE Trans. Pattern Anal. Mach. Intell. 32(Xx), 1–14 (2010)
Spärck Jones, K.: A statistical interpretation of term specificity and its application in retrieval. J. Doc. 28, 11–21 (1972)
Kauppi, T., et al.: The DIARETDB1 diabetic retinopathy database and evaluation protocol. In: BMVC, pp. 1–10 (2007)
Quellec, G., Lamard, M., Josselin, P.M., Cazuguel, G., Cochener, B., Roux, C.: Optimal wavelet transform for the detection of microaneurysms in retina photographs. IEEE Trans. Med. Imaging 27(9), 1230–1241 (2008)
Mizutani, A., Muramatsu, C., Hatanaka, Y., Suemori, S., Hara, T., Fujita, H.: Automated microaneurysm detection method based on double ring filter in retinal fundus images. 7260:72601N–72601N-8 (2009)
Aravind, C., Ponnibala, M., Vijayachitra, S.: Automatic detection of microaneurysms and classification of diabetic retinopathy images using SVM technique. In: IJCA Proceedings on International Conference on Innovations in Intelligent Instrumentation, Optimization and Electrical Sciences ICIIIOES 11, pp. 18–22 (2013)
Ren, F., Cao, P., Li, W., Zhao, D., Zaiane, O.: Ensemble based adaptive over-sampling method for imbalanced data learning in computer aided detection of microaneurysm. Comput. Med. Imaging Graph. 55, 54–67 (2017)
Sehirli, E., Turan, M.K., Dietzel, A.: Automatic detection of microaneurysms in RGB retinal fundus images. Studies 1(8) (2015)
Acknowledgments
This work is supported by National High-tech R&D Program of China (863 Program) (2015AA015904), NSFC (61572316, 61671290, 61525106), National Key R&D Program of China (2016YFC1300302), Key Program for International S&T Cooperation Project (2016YFE0129500) of China, Science and Technology Commission of Shanghai Municipality (16DZ0501100), and Interdisciplinary Program of Shanghai Jiao Tong University (14JCY10).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Dai, L. et al. (2017). Retinal Microaneurysm Detection Using Clinical Report Guided Multi-sieving CNN. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10435. Springer, Cham. https://doi.org/10.1007/978-3-319-66179-7_60
Download citation
DOI: https://doi.org/10.1007/978-3-319-66179-7_60
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66178-0
Online ISBN: 978-3-319-66179-7
eBook Packages: Computer ScienceComputer Science (R0)