Abstract
Protecting crops against insect pests is a major focus area in crop protection. Over the past two decades, biotechnological interventions, especially Bt proteins, have been successfully implemented across the world and have had major impacts on reducing chemical pesticide applications. As insects continue to adapt to insecticides, both chemical and protein-based, new methods, molecules, and modes of action are necessary to provide sustainable solutions. RNA interference (RNAi) has emerged as a significant tool to knock down or alter gene expression profiles in a species-specific manner. In the past decade, there has been intense research on RNAi applications in crop protection. This chapter looks at the current state of knowledge in the field and outlines the methodology, delivery methods, and precautions required in designing targets. Assessing the targeting of specific gene expression is also an important part of a successful RNAi strategy. The current literature on the use of RNAi in major orders of insect pests is reviewed, along with a perspective on the regulatory aspects of the approach. Risk assessment of RNAi would focus on molecular characterization, food/feed risk assessment, and environmental risk assessment. As more RNAi-based products come through regulatory systems, either via direct application or plant expression based, the impact of this approach on crop protection will become clearer.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Gene silencing
- RNA interference (RNAi)
- Insect control
- Double-stranded RNA (dsRNA)
- Short interfering RNA (siRNA)
- Risk assessment
1 Introduction
Crop protection strategies need continuous improvement and innovation since the ability of the insect herbivores to adapt to any pest control intervention is well documented (Georghiou and Lagunes-Tejeda 1991; Storer et al. 2010). The increased strain on agricultural output due to global challenges such as population growth and climate change in conjunction with the escalating costs of chemical pest control, insecticide resistance, and rising environmental and health concerns creates a need for developing new technologies to close yield gaps and minimize environmental impacts (Pradhan et al. 2015). Sustainable intensification of crop production by using the best of conventional plant breeding (adapted germplasm with native resistance) and the best of biotechnology is a theme that is being widely advocated by the global scientific community to meet the daunting challenge of feeding 9.7 billion in 2050 (Smith 2013). New biotechnological techniques developed during the last two decades have helped agriculture to cope with different challenges like pest resistance, disease, herbicide and stress tolerance, and improved yield and product quality characteristics. Transgenic crops expressing Bacillus thuringiensis (Bt) proteins have provided excellent yield protection from insect pest damage, and the success of this technology is evident by the fact that the global planting of crops genetically engineered to express Bt proteins increased to 78 million hectares in 2014 which is a significant fraction of the >170 M ha of transgenic crops cultivated worldwide (James 2014; Baum and Roberts 2014). Although Bt sprays and Bt crops have provided substantial economic and environmental benefits, insect adaptation resulting from the strong selective pressures imposed, has reduced their effectiveness (Tabashnik et al. 2013; Carrière et al. 2015). Therefore, as in the case for synthetic and biological insecticides, alternative modes of action (MOAs) for insect-protected crops are needed, either because some insect species are refractory to Bt proteins or because some have evolved field resistance to the Bt Proteins. To that end, RNA interference (RNAi) which targets and knocks down the expression of genes in a species-specific manner provides significant opportunities for crop protection by managing pest populations and reducing the spread of vector-borne diseases (Price and Gatehouse 2008; Lundgren and Duan 2013). Therefore, there is a growing interest in using dsRNA for insect control, both as a traditional RNAi-based pesticide and RNAi-based genetically modified crop plants.
RNA interference (RNAi) refers to a collection of biological processes, by which exogenously applied and endogenously expressed double-stranded RNAs (dsRNA) target specific endogenous messenger RNAs (mRNAs) for degradation, thereby silencing their expression by making use of conserved cellular machinery (Zamore 2001). Shortly following its discovery in the nematode, Caenorhabditis elegans (Fire et al. 1998), RNAi has been observed in a wide range of eukaryotic organisms and has proved itself to be a powerful tool for investigating gene function (Dykxhoorn and Lieberman 2005). In plants, the dsRNA-triggered sequence-specific RNA degradation pathway has been termed post-transcriptional gene silencing (PTGS). The RNAi pathway is a major antiviral system in plants (Szittya and Burgyan 2013) and nematodes (Sarkies and Miska 2013) and serves as a broadly acting (Kemp et al. 2013) and robust antiviral pathway in insects (Nayak et al. 2013; Bronkhorst and van Rij 2014; Vijayendran et al. 2013). Not only has effective RNAi been demonstrated in many insect species, but it has also been performed in insects in a variety of developmental stages. The evidence for functional RNAi has been reported in a wide range of insect species encompassing different taxonomic groups that include the Coleoptera (Arakane et al. 2004; Suzuki et al. 2008), Diptera (Lum et al. 2003; Dietzl et al. 2007), Dictyoptera, Hemiptera, Hymenoptera (Schluns and Crozier 2007; Antonio et al. 2008), Isoptera, Lepidoptera (Chen et al. 2008; YuQ et al. 2008; Tian et al. 2009; Terenius et al. 2011), Neuroptera, and Orthoptera. Recent studies have shown the potential applications of this tool in fundamental and applied research and more specifically for crop protection against insect pests. The ingestion of double-stranded RNAs targeting essential insect genes by insects can trigger RNAi and lead to growth inhibition, developmental aberrations, reduced fecundity, and mortality. RNAi can therefore be considered as one of a suite of tools for crop improvement and insect protection.
The range of potential applications of RNAi in agriculture is remarkable, and the technology is being evaluated to introduce novel plant traits, increase crop yield, and improve product quality. RNAi is being touted as the “game changer” in agriculture because it has provided a highly specific, non-chemical solution for pest and pathogen control (Baum et al. 2007; Price and Gatehouse 2008; Huvenne and Smagghe 2010). The remarkable systemic nature of this mechanism in insects makes RNAi-based insecticides an exciting new IPM alternative for agricultural pest control (Price and Gatehouse 2008). Proof-of-concept studies clearly illustrate the efficacy of this technology for crop protection. RNAi-based transgenic plants intended for market release can be designed to either induce silencing of target genes in planta or in insect pests (Koch and Kogel 2014). RNAi-based GM crops have been developed in the laboratory for three major crops: corn (Baum et al. 2007), cotton (Mao et al. 2007; Mao et al. 2011), and rice (Zha et al. 2011). Although RNAi was perceived to provide greater specificity in pest control and have little or no off-target effects, many studies have shown that unintentional off-target gene silencing in target cells and gene silencing in non-target organisms occur more commonly than expected (Baum et al. 2007; Qiu et al. 2005; Mohr and Perrimon 2012). An overall picture of the field risks, the environmental fate of dsRNA and RNAi effects in variation trials (differential effects of RNAi treatments), is emerging (Chu et al. 2014; Palli 2014; Lundgren and Duan 2013). Our review focuses on the current knowledge of RNAi mechanism in general and highlights the scientific data generated in insects with respect to mechanism, dsRNA uptake, and how RNAi can complement the existing technologies for crop protection for achieving optimum crop productivity.
2 RNAi Mechanisms and Machinery
Important insights have been gained in elucidating the detailed mechanism of RNAi, since its initial discovery, and definitions have been proposed to differentiate the various aspects of RNAi in plants and animals. Whangbo and Hunter (2008) categorized the RNAi response into the following three types: cell autonomous, environmental, and systemic. Environmental and systemic RNAi are together referred to as non-cell autonomous RNAi. Cell autonomous RNAi refers to the silencing effect that is encompassed within the cells where dsRNA is constitutively expressed or exogenously introduced. In the non-cell autonomous RNAi, the silencing signal is directly picked up by cells from the environment, viz., gut or hemocoel. The phenomena in which the silencing signal (siRNA and/or dsRNA) spreads to neighboring cells or remote tissues from an epicenter of cells is called systemic RNAi, whereas in environmental RNAi, RNAi pathway is triggered by environmental exposure (either by soaking or feeding), and this may or may not be followed by systemic movement of the silencing signal (Baum and Roberts 2014). The presence of non-cell autonomous RNAi in arthropods and its specificity are two important factors that paved the way for using this technology in pest control. However, for the successful application of the technology as a crop protection agent, environmental RNAi must first be evaluated, and a suitable delivery system for dsRNA has to be identified.
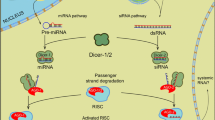
The RNAi machinery can be categorized into two functional groups: (1) the intracellular machinery, consisting of the Dicer and Argonaute proteins, and (2) the “systemic machinery,” composed of factors that amplify the dsRNA trigger and allow it to spread to other tissues within the animal or even to the next generation (Siomi and Siomi 2009; Swevers 2012). The RNAi pathway is initiated upon recognition of the long dsRNA precursor molecule that varies in length and origin and can be introduced into the cell through microinjection, transfection, or expression from endogenous genes (Huvenne and Smagghe 2010; Whangbo and Hunter 2008). dsRNA can also move into the cells through the transmembrane transporters SID1/2 or the endocytosis machinery (Feinberg and Hunter 2003; Jose and Hunter 2007). The dsRNA precursors are processed by Dicer-2, a ribonuclease III (RNase III) family dsRNA endonuclease, into ~ 19–25 nt long siRNA duplexes with characteristic 2 nucleotide (nt) 3′ overhangs. The dsRNA-binding motif proteins (dsRBMs) facilitate the assembly of the siRNAs with the RNase H enzyme Argonaute-2 (Ago2), to form a multi-protein RNA-induced silencing complex (RISC) (Hammond et al. 2001), where one of the siRNA strands (the passenger) is degraded in a process dependent upon Ago2 and the endoribonuclease C3PO (component 3 promoter of RISC) (Liu et al. 2009). The other strand (the guide) is retained and remains associated with Ago2 and is 2′-0-methylated on its 3′ terminal nt by the Hen1 methyltransferase, thus creating a mature RISC (Horwich et al. 2007; Saito et al. 2007). Base pairing of the guide strand to a complementary target single-stranded RNA (including mRNAs) leads to Ago2-mediated degradation of the target (Meister and Tuschl 2004). The siRNA mechanism of the RNAi pathways is harnessed as an experimental tool to target and degrade specific mRNAs with sequence homology to the administered/incorporated dsRNA molecules.
The fundamental components of the RNAi machinery are evolutionarily conserved among insects and are readily identified in insect species whose genomes have been sequenced (Zhu et al. 2014). The ribonuclease III enzyme Dicer, one of the key enzymes involved in RNAi pathways, is encoded by variable number of genes and presents distinct functions among organisms. While mammals and nematodes have a single Dicer responsible for functions in siRNA and miRNA pathways (Ghildiyal and Zamore 2009), insects have two Dicer proteins, Dcr-1 and Dcr-2 that are assigned to the miRNA and siRNA pathways (Lee et al. 2004). Dcr-1 preferentially processes the pre-miRNA to miRNA, whereas Dcr-2 is in charge of processing long dsRNA into siRNAs (Tomoyasu et al. 2008; Aronstein et al. 2011; Asgari 2013). miRNAs are processed from endogenous genes and function in the regulation of gene expression, while the siRNAs are derived from dsRNA molecules and provide defense against invading viruses. The argonaute family proteins (AGO) are the central protein components of the silencing complexes (RISC) that act in mediating target recognition and silencing (Peters and Meister 2007) and have been observed in different insect taxonomic groups (Aronstein et al. 2011; Swevers et al. 2013). The Ago proteins with a proven role in determining RNAi efficiency were found to be duplicated in the Tribolium castaneum genome (Tc-Ago-2a and Tc-Ago-2b), whereas Drosophila carries only one copy of the AGO-2 gene, thereby suggesting a relationship between number of copies of AGO gene and insect RNAi response (Tomoyasu et al. 2008).
In animals, the uptake of dsRNA is facilitated by two machineries, viz., the transmembrane channel-mediated uptake machinery based on SID-1(systemic interference defective-1) and SID-2 proteins (Jose and Hunter 2007) and the endocytosis-mediated uptake machinery (Saleh et al. 2006). In C. elegans, the SID-1 protein is inferred to function as a dsRNA channel (Winston et al. 2002), and the SID-2 has been implicated in dsRNA uptake by gut cells and probably functions in environmental RNAi (Jose and Hunter 2007). SID-1 homologues are detected in the genomes of insects belonging to Coleoptera, Lepidoptera, Hymenoptera, and Hemiptera but not Diptera (Gordon and Waterhouse 2007). Although putative insect orthologs of the C. elegans sid genes have been described in various insect species, their involvement in RNAi is still not known (Xu and Han 2008; Huvenne and Smagghe 2010). The sid-1-like genes of insects show greater sequence homology with the C. elegans gene chup-1, a cholesterol transporter that has no involvement in RNAi, than to sid-1 (Valdes et al. 2012; Luo et al. 2012). In Drosophila melanogaster, which lack a SID gene ortholog, dsRNA uptake occurs by receptor-mediated endocytosis (Saleh et al. 2006). In the nematode C. elegans and in many plants, there exists a host-derived RNA-dependent RNA polymerase (RdRp), for amplifying the silencing signals by generating “secondary siRNAs” that sustain the RNAi response (Carthew and Sontheimer 2009). There is no evidence of such RdRp homologue in any insect genome sequenced to date (Tomoyasu et al. 2008). There is considerable ambiguity on how the RNAi triggered by the acquisition of dsRNA molecules is sustained in insect cells, and it is speculated that some unknown mechanism may be responsible for the systemic RNAi response (Barnard et al. 2012).
3 Important Considerations of an RNAi Experiment
Although RNAi is a highly conserved cellular mechanism and its use for the control of insect herbivores seems to be very straightforward, RNAi application and efficacy remain variable between genes, life stages, and organisms. Factors that determine the success of RNAi experiments in different insect species include the uptake of dsRNA (environmental RNAi and/or systemic RNAi), presence/absence of the core RNAi machinery, cellular uptake and propagation of signal (Roignant et al. 2003; Miller et al. 2008), and dsRNA degrading enzymes (Arimatsu et al. 2007), as well as other differences in genetic backgrounds (Kitzmann et al. 2013; Scott et al. 2013). These biological variables have been experimentally studied in different insect species. A detailed description of different factors influencing the success of RNAi technology in insects is presented below.
3.1 Identification of Target Genes
For successful RNAi, the choice of the essential genes that can trigger a lethal RNAi response in the insect pest requires careful consideration. Target gene selection is crucial yet challenging especially for those pest species that are usually difficult to rear in the lab or those that lack the required genomic and genetic tools for a whole animal-high-throughput-screen. In such cases, data from appropriate insect model systems can be subjected to large scale unbiased RNAi screens for target gene identification (Ulrich et al. 2015). The abundance of target gene transcript and the rate of protein turnover are two important factors that influence the outcome of an RNAi experiment. An mRNA pool with high turnover that codes for a protein with a short half-life is considered an ideal gene target for RNAi (Scott et al. 2013). Phenotypic evaluation of gene function using RNAi may not be easy for a stable protein with a long half-life. Another limitation is that for majority of genes, mRNA turnover and protein half-life are not known. Potential target genes which will function under field conditions can be identified by performing bioassays that closely mimic the conditions in the field. A precise choice of the target region from the target gene for dsRNA synthesis will ensure the specificity of RNAi and concurrently limits the off-target effects. Identification of essential targets is possible by extended literature search, analyses of available DNA/RNA sequence databases, and gene screening mediated by second-generation sequencing (Andrade and Hunter 2016; Wang et al. 2011). Insect genomics research initiatives have steadily increased in the last two decades due to the availability of cost-effective, high-throughput DNA sequencing platforms that have contributed to the sequencing of genomes of agriculturally important organisms. These efforts have broad implications in insect functional genomics studies, enhancing the throughput of RNAi target identification and development of insect management technologies.
3.2 Designing the RNAi Molecule
When designing RNAi experiments, important considerations regarding the design of a specific RNAi molecule (in the form of dsRNA, siRNA, or a hairpin RNA) for a target gene of interest (GOI) include the length of the molecule, sequence identity to the target transcript of the insect, and the region targeted within the mRNA (Scott et al. 2013; Andrade and Hunter 2016). The length of dsRNA is an important parameter for successful RNAi as different efficacies by different sizes of dsRNA were reported by Whyard et al. (2009) and Saleh et al. (2006). Although the minimal required length to achieve an optimum RNAi effect varies among insect species (Bolognesi et al. 2012), greater success with insect RNAi has been achieved with dsRNA molecules of ≥50–200 bp in length (Huvenne and Smagghe 2010). Huvenne and Smagghe (2010) provide a comprehensive survey of the length range of dsRNAs used in early studies: from 134 to 1842 bp, with most studies using 300–520 bp. The advantages of using longer >200 bp dsRNA for RNAi strategies in pest management are the production of many siRNAs against the targeted mRNA transcript, potentially maximizing the RNAi response. Occasionally, designing of RNAi molecules, shorter in length than ideal, may achieve desired specificity. Studies have shown the effectiveness of chemically synthesized siRNAs (obtained by dicing the dsRNA in vitro before delivery to the insect) in the suppression of target gene expression.
Another important aspect in the design of dsRNA sequences is the stringency to be adopted in order to achieve the desired specificity and avoid/minimize off-target effects. This property facilitates the designing of species-specific sequences that mediate insect lethality and is a prerequisite for taking the technology from the laboratory to the field. As reported by Zhang et al. (2010), two genes with high sequence similarities can both be silenced by the same dsRNA. This has important implications in off-target effects. Therefore the target gene and target region should be carefully determined in order for adequate and specific RNAi. The gene regions (e.g., 5′ or 3′ end of the gene) to which RNAi molecules are designed have also yielded variable results, thus emphasizing the importance of screening multiple RNAi sequences for a gene of interest (Mao and Zeng 2012; Pridgeon et al. 2008; Loy et al. 2012).
The RNAi molecule design process can be aided by software tools, algorithms, and databases that evaluate the genome sequence and RNA folding kinetics to optimize effectiveness. Following are some of the online software tools that are available to minimize off-target effects and achieve specific and precise silencing effect: the NEXT-RNAi software enables the design and evaluation of siRNAs and long ds-RNAs and can be used for the design and evaluation of genome-wide RNAi libraries in an organism-independent manner for all sequenced and annotated genomes. The input data for the analysis are the desired target sequences and an off-target database. The Next-RNAi software was deployed to design novel genome-wide RNAi libraries of long dsRNA for the following insects, viz., D. melanogaster, T. castaneum, and Anopheles gambiae, and to design multiple RNAi for a specific gene to study associated phenotype (Horn et al. 2010). The web-based E-RNAi tool initially developed for RNAi experiments in C. elegans and Drosophila (Zeynep et al. 2005) provides siRNA and long dsRNA design suggestions suitable for RNAi experiments in a variety of other species and insects that include Apis mellifera, T. castaneum, Acyrthosiphon pisum, A. gambiae, and Aedes aegypti (Horn and Boutros 2010). It can calculate off-target impacts that may affect the phenotypic results. The dsRNA sequences are evaluated for their specificity and efficiency. The dicer enzyme in the RNAi machinery cleaves long dsRNA into small 19–22 nucleotides long siRNAs. dsCheck is a software that investigates individual 19 nucleotide fragments of long dsRNA and produces a list of potential off-target gene candidates based on its novel algorithm. This tool provides off-target search to verify previously designed dsRNA sequences and also presents “off-target minimized” dsRNA design (Naito et al. 2005).
3.3 Delivery of dsRNA for Insect RNAi
The effective introduction of the RNAi trigger into an organism and its subsequent entry into the RNAi pathway is the most limiting factor of the RNAi experiment. There are many methods of dsRNA delivery reported and applied in RNAi experiments, including microinjection (Tan et al. 2008; Martin et al. 2006), feeding (in vitro synthesized dsRNA) (Zhou et al. 2008; Zhu et al. 2011), transgenic plants expressing dsRNA(Baum et al. 2007; Mao et al. 2007), nanoparticle RNAi (Zhang et al. 2010), soaking (Terenius et al. 2011; Ulvila et al. 2006), and topical application (Pridgeon et al. 2008). Intracellular RNAi results from the expression of hairpin RNAs as transgenes or during the introduction of dsRNA into cells by electroporation or transfection or by direct delivery into a cell. Extracellular RNA, which requires the uptake of dsRNA molecules by the cells, is achieved by soaking, feeding, or injection into the hemocoel (Yu et al. 2013). From the studies it is evident that most insect RNAi studies relied on the delivery of specific dsRNA triggers through either microinjections (Fire et al. 1998; Adams et al. 2000) or ingestion through feeding (Ulvila et al. 2006; Rangasamy and Siegfried 2012). Each of these methods has its own advantages and limitations which are discussed further.
3.3.1 Delivery of dsRNA Trigger Through Injection (Microinjection)
Microinjection is a widely used dsRNA delivery method in arthropods, and the first successful microinjection experiment performed in vivo was in D. melanogaster embryos in which the expression of frizzled and frizzled 2 genes was down regulated by intracellular RNAi (Kennerdell and Carthew 1998). This technique was subsequently used to deliver dsRNA to the giant silk moth, Hyalophora cecropia (Bettencourt et al. 2002). Although, the sequencing of D. melanogaster genome in 2000 (Adams et al. 2000) made RNAi a popular research tool in functional genomics of this model insect, extracellular RNAi seems to have limited application in this species (Dzitoyeva et al. 2001). A plausible reason for this could be the fact that the cells in most larval tissues seem to be recalcitrant to the uptake of dsRNA from outside the cell (Miller et al. 2008), whereas some tissues in the adults are able to take up dsRNA (Tomoyasu et al. 2008), thus limiting the use of RNAi in gene function studies. In T. castaneum, a stored product pest, and a coleopteran model insect, microinjection of dsRNA, both in larvae and adults, is widely used in functional genomics studies, as this species shows a robust systemic RNAi response (Tomoyasu and Denell 2004). Comprehensive microinjection protocols for RNAi experiments have been published for the two model insects, Tribolium and Drosophila, and these protocols provide a quick reference and standard for similar experiments in other arthropod species. Successful delivery of dsRNA by injection has also been demonstrated in Lepidoptera; however, the method has shown great variation in effectiveness between species and is not as simple as shown in other taxa (Terenius et al. 2011; Kolliopoulou and Swevers 2014). Nonetheless, quite a number of RNAi microinjection experiments have been performed in species from the order Lepidoptera with most notable success achieved with Bombyx mori and Manduca sexta, and the members of the Saturniidae family were found to be quite sensitive to RNAi using hemocoel injection as the dsRNA delivery method, compared to other species within the order (Yu et al. 2013). In the moth species, RNAi based on dsRNA microinjection has been applied to all life stages, viz., egg (Osanai-Futahashi et al. 2016; Fabrick et al. 2004), larvae (Mohammed et al. 2015; Sun et al. 2016; Zhao et al. 2013), pupa (Choi et al. 2012; Qian et al. 2015), and adults (Abrieux et al. 2013; Hassanien et al. 2014). Injection of dsRNA has been proven successful to cause a knockdown effect in the economically important model insect, the A. mellifera (Farooqui et al. 2003; Gatehouse et al. 2004; Aronstein and Saldivar 2005).
Microinjection has also been used to deliver dsRNA or siRNA for RNAi in the agriculturally important hemipteran herbivores, the pea aphid, A. pisum (Jaubert-Possamai et al. 2007; Mutti et al. 2006), whitefly, Bemisia tabaci (Ghanim et al. 2007), and nymphs and adults of the small brown plant hopper, Laodelphax striatellus (Liu et al. 2010). Microinjection of long dsRNA into the body cavity of B. tabaci caused downregulation of genes uniquely expressed in the midgut and salivary glands, and injection of dsRNA targeting the whitefly Drosophila chickadee homologue caused phenotypic effects in the ovaries of B. tabaci. The disruption of gene expression in the hemipteran herbivores opens the door to new strategies aimed at curbing down the deleterious effects of these insect pests to agriculture (Ghanim et al. 2007).
Microinjection protocols are currently available for various taxa, including Lepidoptera, Diptera, Hymenoptera, the Orthopterans, and Cockroaches (Terenius et al. 2011; Blandin et al. 2002; Martin et al. 2006; Belles 2010; Huang and Lee 2011; Nakamura et al. 2008). Microinjection has been applied to all life stages in hemi- and holometabolous insects, and a large variation in the success rates of these experiments has been observed between different species, genera, and taxa (Yu et al. 2013). In the case of larvae, injections are usually carried out dorsally or between segments, whereas in adults the tissue under the wings is the easiest location to inject the organism. Microinjection has both its advantages and disadvantages compared to the other methods of dsRNA delivery. This technique allows researchers to get the dsRNA directly and effectively into the tissue of choice or into the hemolymph without being hindered by barriers such as the integument or the gut epithelium, in addition, to providing the flexibility to deliver the precise amount of dsRNA. However, an important shortcoming of this technique in insects is the mechanical damage during the injection, which is quite significant when targeting embryos and neonatal larvae and pupae (Scott et al. 2013; Yu et al. 2013). The mechanical damage may also have undesirable effects or even obscure the targeted effects especially when studying the function of genes relation to behavior and survival using RNAi. Furthermore, this method is time-consuming and labor-intensive, requires expertise, can only be used in the laboratory, and is not suitable for RNAi-based pest control (Xu et al. 2016).
3.3.2 Delivery of dsRNA Trigger Through Ingestion
Feeding is another method for introducing dsRNA into an organism for triggering RNAi. RNAi triggered by ingested dsRNA was first demonstrated in C. elegans (Timmon and Fire 1998; Timmons et al. 2001) and subsequently applied in various insects and taxa such as Spodoptera exigua, Diabrotica virgifera virgifera, and Epiphyas postvittana (Turner et al. 2006; Baum et al. 2007; Tian et al. 2009; Surakasi et al. 2011). The dsRNA used for ingestion experiments can either be expressed in bacteria or plants, or they can be synthesized in vitro and then fed to insects either by mixing with food or by supplying as solution droplets. Uptake of bacterially expressed dsRNA was applied in S. exigua to suppress the expression of S. exigua chitin synthase A (SeCHSA) gene, a non-midgut gene specifically expressed in the cuticle and trachea of S. exigua (Tian et al. 2009). The study further established that the phenotypes recovered post-ingestion were dependent on the dsRNA dosage and accumulation. Transcriptional suppression of target gene expression in salivary glands of the tick, Ixodes scapularis (Soares et al. 2005), and fat body tissue of Reticulitermes flavipes (Ulvila et al. 2006) was observed when dsRNA was delivered through ingestion route. The oral delivery of in vitro synthesized dsRNA either by dissolving the dsRNA in liquid artificial diets (Sadeghi et al. 2009) or overlaying on the surface of solid foods was used in T. castaneum, A. pisum, and M. sexta for the knockdown of a species-specific E-subunit of the vATPase gene (Whyard et al. 2009), which led to 50–75% mortality in all three insect species (Yu et al. 2013). RNAi triggered by ingested dsRNA that was delivered via artificial diet surface coated with dsRNA or food that was mixed with dsRNA was effective in Drosophila species (Whyard et al. 2009). Transient suppression of β1 integrin subunit (βSe1) expression in S. exigua gut epithelium was achieved by providing dsRNA-treated cabbage leaf disks to fourth instar larvae, and significant mortality was also recorded (Surakasi et al. 2011).
The dsRNA droplet feeding as described by Turner et al. (2006) is yet another method applied in the research on a larval gut carboxylesterase gene (EposCXE1) and the adult antennae-expressed pheromone binding protein (EposPBP1) gene in E. postvittana larvae and a cytochrome P450 (CYP6BG1) gene in Plutella xylostella (Bautista et al. 2009). Successful knockdown of the target gene and significant RNAi effects were observed in these studies proving the efficacy of ingestion as a method for effective RNAi. Nanoparticle-mediated RNAi technique, in which dsRNAs were entrapped by the polymer chitosan via electrostatic forces to form a chitosan/dsRNA nanoparticle, was another innovation for delivering the dsRNA to insect by ingestion (Zhang et al. 2010). Formation of nanoparticles was believed to enhance the efficacy of RNAi by providing improved stability to the dsRNA molecule through the delivery process (Yu et al. 2013). Oral delivery of dsRNA can also be achieved by exposing the target insects to transgenic plants that express hairpin dsRNAs targeting specific genes from insects to increase their resistance to herbivorous insects (Baum et al. 2007; Mao et al. 2007). Silencing of genes in target insects of Lepidoptera, Coleoptera, and Hemiptera was evaluated effectively by delivery of dsRNAs through transgenic plants (Baum et al. 2007; Pitino et al. 2011; Zha et al. 2011). Model plants such as thale cress (Arabidopsis thaliana) (Zha et al. 2011; Liu et al. 2015), tobacco (Nicotiana tabacum) (Mao et al. 2007), rice (Oryza sativa) (Zha et al. 2011), tomato (Solanum lycopersicum) (Mamta and Rajam 2016), and cotton (Gossypium hirsutum) (Mao et al. 2011) were transformed to express dsRNA against target herbivores.
Oral delivery of dsRNA into insects for RNAi can thus be performed by any of the following approaches, viz., artificial diet, detached plant parts, or intact plants, and provides several advantages. Not only is this technique easy to perform, it is a labor-saving, cost-effective, and comparatively less invasive method with the potential for high-throughput screening of target genes and potential for field application (Tian et al. 2009; Kamath et al. 2000). This method may be the most suitable method for developing RNAi pesticides since it allows RNAi through pest insect feeding on sprayed dsRNA-based pesticide or transgenic plant and bacteria that express dsRNA (Xu et al. 2016). The limitations of oral delivery of dsRNA include the limited or no efficiency of dsRNA ingestion in inducing RNAi in some insect species, thereby suggesting that the technique may not be suitable for all species. In S. litura, ingested dsRNA targeting a gut-specific aminopeptidase N failed to induce RNAi (Rajagopal et al. 2002). The gut environment of the target insect species and the final effective dosage/concentration delivered or needed for RNAi are difficult to determine and optimize, which could compromise the investigations (Turner et al. 2006; Surakasi et al. 2011).
3.3.3 Delivery of dsRNA Trigger Through Soaking and Transfection
Soaking, as a method for successful delivery of dsRNA and induction of specific RNAi response, was first reported in C. elegans (Tabara et al. 1998) and subsequently applied to large-scale analysis of gene function in nematodes and other species. This method is particularly suitable for RNAi analysis in insect cell lines and tissues, as well as in specific life stages of insects, such as eggs and neonate larvae (Yu et al. 2013; Wang et al. 2011; Wu et al. 2016; Singh et al. 2013). The S2 cells derived from D. melanogaster embryos were used for initial soaking experiments by adding specific dsRNA to the cell growth medium to suppress specific gene expression, and then subsequently this method became the most commonly used method for inducing RNAi response in S2 cells (Clemens et al. 2000; Caplen et al. 2000; Shah and Förstemann 2008). Since then, RNAi experiments in Sf21 cells derived from ovaries of S. frugiperda were conducted using this technique, in which the downregulation of target genes was accomplished by soaking the cells in dsRNA (Sivakumar et al. 2007) and siRNA (Agrawal et al. 2004) solutions. It was however observed that simple soaking of the cells in dsRNA supplemented culture medium was not sufficient to trigger an RNAi response in some species, but introduction of the dsRNA into the cells by transfection was probably more efficient technique when compared to soaking (Beck and Strand 2005; Valdes et al. 2003). Soaking appears to work with similar efficiency as feeding in C. elegans; however, it is not as effective as microinjection (Tabara et al. 1998). The method is easy to use and is suitable for conducting high-throughput RNAi screens (Perrimon and Mathey-Prevot 2007) and genome-wide analysis in the study of phenotypes characterization (Sugimoto 2004).
3.3.4 Other dsRNA Delivery Methods
Other methods to introduce dsRNA trigger into organisms, including electroporation (Osanai-Futahashi et al. 2016), virus-mediated delivery (Kontogiannatos et al. 2013), and bacterial symbiont mediated delivery (Whitten et al. 2016), have also been applied to RNAi in insects including moths (Xu et al. 2016).
3.4 Impact of dsRNA Dosage on RNAi
Optimal concentration of dsRNA uptake by an insect determines the final outcome of RNAi, and the dose required varies with insect species, developmental stage of the insect, the abundance of target gene transcript, and the delivery method used. A higher dose is usually required when the RNAi molecule is delivered orally as compared to injection. For RNAi studies using microinjection, it is not possible to use very high concentrations because of the viscous nature of the dsRNA solutions. Although very high doses of dsRNA can be incorporated into the insect artificial diets, synthesizing large amounts of dsRNA may not be cost-effective, thereby presenting a major experimental bottleneck (Scott et al. 2013). The presence of nucleases in various extracellular fluids of insects, such as the saliva of Lygus lineolaris (Allen and Walker 2012), the digestive juices of B. mori (Arimatsu et al. 2007), and the hemolymph of M. sexta (Garbutt et al. 2013), also impacts the dosage of RNAi trigger that can cause effective RNAi as the breakdown of dsRNA may not provoke an optimum RNAi response. In the pea aphid, A. pisum, a lack of response in RNAi feeding and injection assays was associated with the degradation of dsRNA by both the salivary secretions and the hemolymph (Christiaens et al. 2014). Finally, insect gut pH is yet another factor that influences the efficiency of RNAi. It has been observed that gut pH is quite variable among insect orders with the pH being predominant acidic in Coleopteran larvae to strongly alkaline in some species of Lepidoptera. All of these biochemical factors in the gut greatly influence the stability of dsRNA and impact the bioavailability of dsRNA to suppress target gene expression (Price and Gatehouse 2008). The strong alkaline gut pH in the Lepidopteran larvae may be a contributing factor in the species from this order for being recalcitrant to gene silencing by RNAi. The effects of RNAi in insects usually begin to appear within 4 to 5 days post-ingestion suggesting that there may be a dose response (Yu et al. 2013). In several insects, the RNAi efficiency is either low or variable at best, and therefore a relatively high dosage of RNAi trigger molecules is needed to compensate for the species- and tissue-specific antagonistic biological factors, viz., degradation of dsRNA and weak activity of the RNAi machinery that impact the efficacy of RNAi. Furthermore, the mode of uptake, the ability to process RNAi molecules, and the ability to spread the signal are other important factors that influence the essential dose required to induce an RNAi response.
3.5 Evaluation of RNAi Experiments
The desired result of an RNAi experiment varies with the objective of the study (gene function analysis or insect control). For research attempting to develop novel RNAi-based product for insect control, a successful outcome would be to obtain high insect mortality, whereas for gene function analysis, physiological indices of predicted function should be central to the analysis. Therefore, defining and integrating the appropriate physiological and fitness assays in the experimental design are critical (Scott et al. 2013). Furthermore, in order to expedite the process of development of a viable pest control product using RNAi strategy, identifying the best delivery mechanism (i.e., topical sprays, baits, or transgenic plants) early in the product development phase is of utmost importance (Andrade and Hunter 2016).
3.5.1 Bioassays
RNAi bioassays for insects are optimized taking into consideration the feeding behaviors of insects, and the in vitro experiments are designed to mimic conditions the insects will encounter in the field. A range of concentrations of dsRNA are tested to select an optimal concentration for effective RNAi. For insects with piercing-sucking mouthparts, artificial feeding bioassays are being used widely for RNAi. However, major drawbacks associated with liquid feeding bioassays (dsRNAs mixed in a liquid diet or a sucrose solution) include the high mortality levels observed in the controls, the significantly higher dsRNA concentrations required to achieve mortality, and the increased degradation of dsRNA in the liquid diet due to bacterial or fungal contaminations (Upadhyay et al. 2011). Also, it has been reported that concentrations of up to 1 μg/μL cannot be reproduced inside plant vascular tissues (Borgio 2010; Katoch et al. 2013; Tomizawa and Noda 2013), which is an important parameter to consider when developing an effective RNAi control strategy against the plant sap feeders (Andrade and Hunter 2016). Successful oral uptake of dsRNA delivered via host plants treated with dsRNA either as a foliar spray or root drench was demonstrated in two hemipteran insects, the xylem-feeding leafhopper (Homalodisca vitripennis) and the phloem-feeding Asian citrus psyllid (Diaphorina citri) (Hunter et al. 2012). Cost-effective feeding bioassays for screening large number of dsRNA molecules against the hemipteran pests can be developed using leaf disks, whole leaf, new growth leaves and stem, or root cuttings (Andrade and Hunter 2016). These bioassays can be terminated after 8–10 days of observations for mortality and may be extended further to record observations on insect oviposition, egg viability, or nymph development, since the plant material was found to remain viable for up to 40 days on an average. These bioassays provide the flexibility to screen for the synergistic effects of multiple dsRNAs in addition to screening single dsRNA against multiple insect herbivores of a specific host plant (Andrade and Hunter 2016). For bioassays in chewing insects, viz., the Lepidoptera and Coleoptera, which are foliage feeders, topical foliar spray is a suitable method for delivery of dsRNA. Host plant leaves sprayed with dsRNA solution are fed to the insect, and the RNAi effects are evaluated. The effectiveness of this procedure was reported with the Coleopteran insects such as western corn rootworm (WCR) (Bolognesi et al. 2012), Colorado potato beetle (CPB) (Miguel and Scott 2016), the diaprepes root weevil (DRW), and Diaprepes abbreviatus L. (Andrade and Hunter 2016) and in several other species as well. In WCR, the dsRNA delivered as a foliar spray silenced genes in tissues far from the gut epithelium, and in CPB, the actin dsRNA conferred protection against insect damage for at least 28 days under greenhouse conditions, and the dsRNA was found to be quite stable. Since chewing insects tend to consume a lot of leaf material each day, a low-dose spray may be able to deliver a significant amount of RNAi trigger.
3.5.2 Controls
When conducting RNAi experiments, depending on the type of assay/treatment, including a negative control, viz., empty vector, empty cassette, buffer only, or a non-specific control (such as dsGFP (green fluorescent protein) gene region), is essential as this will aid in discriminating specific gene silencing from the simple induction of siRNA processing machinery by exposure to a dsRNA. The reporter dsRNA used as a negative control is selected on the basis of having no off-target effects; hence, it should not show sequence similarity to any known insect mRNA transcript.
3.5.3 Quantifying the Transcript Levels
The most commonly used method for tracking effectiveness of RNAi is the RT-qPCR, a method that measures the successful reduction of transcript levels as a result of RNAi and expressed as a percent reduction of the relevant transcript in the treatment group versus the negative control group. Although this method is widely used and accepted, the choice of the reference or housekeeping genes for calculating relative transcript levels is challenging, and these genes although identified and validated on the species level show variable expression depending on the physiology of the insect and the tissue being targeted.
The final phenotypes obtained as a result of a successful RNAi experiment depend on the reduction of protein levels for the gene of interest; therefore, it is highly desirable to determine relative protein concentration. However, there is a possibility of no correlation between the protein levels and the level of transcript suppression; in addition a distinct phenotype may not be observed or recovered despite successful suppression of transcript levels especially when redundancy is built into a specific biological function (Scott et al. 2013). Proteins with a long half-life interfere with the phenotypic changes. For example, a significant phenotypic change in spinosad sensitivity was not observed when RNAi was used to suppress the expression of an α6 nicotinic acetylcholine receptor subunit involved in spinosad toxicity in both D. melanogaster and T. castaneum, suggesting that the RNAi may not be an appropriate method to study the role of target genes where the protein is stable for longer periods (Rinkevich and Scott 2013).
4 Insecticidal RNAi and Crop Protection
The potential of insecticidal RNAi for crop protection and management of insect herbivores and beneficial insects is widely recognized, and the following options, viz., transgenic plants expressing the insecticidal RNAi trait (plant-mediated RNAi) and using dsRNA as a traditionally applied insecticide, are being pursued by industry and academia for product development (Xue et al. 2012; Lundgren and Duan 2013). RNAi offers exquisite specificity and flexibility that cannot be matched by other crop protection interventions such as chemical insecticides, biological control, or protein-coding transgenes (Scott et al. 2013). The breakthrough in applying insecticidal RNAi strategy for controlling agricultural pests via transgenic plants expressing hairpin dsRNA to target specific gene regions of the insect pests came from studies conducted on the western corn rootworm, D. v. virgifera (WCR) (Baum et al. 2007), and cotton bollworm, Helicoverpa armigera (CBW) (Mao et al. 2007). Baum et al. (2007) screened 290 gene targets for evaluating RNAi response in larval WCR, from which they observed rootworm mortality or stunting in approximately 2/5 of the targets screened at concentrations as low as ~50 ng/cm2 in surface overlay diet bioassays (Baum et al. 2007; Baum et al. 2011). A distinct phenotypic response was not observed on suppression of certain gene targets (Baum and Roberts 2014). A gossypol-induced cytochrome P450 gene, CYP6AE14, was targeted for RNAi analysis in CBW by Mao et al. (2007). The gene is expressed in the larval midgut and permits the bollworm to tolerate inhibitory concentration of the cotton secondary metabolite, gossypol. When CBW larvae were fed A. thaliana or N. tabacum leaves expressing CYP6AE14 dsRNA, lower expression levels of this transcript was observed in the midgut, and larval growth was retarded, and both effects were more dramatic in the presence of gossypol. The demonstration of plant resistance to insects mediated by the RNAi-based trait has not only added a new tool to the crop protection toolkit but has exemplified the following key issues for successful environmental RNAi in crop pests: a large number of specific gene targets are available that can be screened, the choice of the target sequence(s), the size of the RNAi trigger, and the mode of delivery of the RNAi trigger (Baum et al. 2007; Mao et al. 2007; Bolognesi et al. 2012; Khajuria et al. 2013).
Due to the long product development timelines, slow regulatory approval of plant delivered RNAi, and the recalcitrant nature of species in some taxa toward environmental RNAi, the future of the RNAi-based insect pest management strategies may depend on non-transformative RNAi strategies and development of topical formulations (Hunter et al. 2010; Baum and Roberts 2014). Alternative approaches are being developed for using RNAi strategy as a conventional topically applied pesticide. The use of topical sprays relies on the penetration or adsorption of the RNAi trigger through the insect cuticle, thereby bypassing the insect gut (Wang et al. 2011). For achieving commercial success with the RNAi strategy as a conventional topically applied pesticide, efficient methods for production, delivery, and increased stability of dsRNA have to be developed. Nanoparticle-mediated RNAi was found to be an effective delivery method and provided a better stability of dsRNA in some insects (Yu et al. 2013; Zhang et al. 2010; Palli 2014). The delivery of dsRNA using nanoparticles reduces dsRNA degradation and increases cellular uptake of intact dsRNA (Joga et al. 2016). The perceived advantages in using chitosan nanoparticles such as being low-cost, enabling high-throughput evaluation of phenotypes (Mysore et al. 2014), and being nontoxic besides their biodegradable nature (Dass and Choong 2008) make them a novel tool for dsRNA delivery (Zhang et al. 2010). Chitosan nanoparticles were used to demonstrate gene knockdown effects in A. gambiae (Zhang et al. 2015) and diet-based delivery of chitosan nanoparticles suppressed gene expression in Asian corn borer (He et al. 2013). Recently, topical application of pathogen-specific dsRNA for virus resistance in plants was reported (Mitter et al. 2017). dsRNA was loaded on designer, nontoxic degradable, layered double hydroxide (LDH) clay nanosheets, and the complex is referred to as “BioClay.” BioClay offered protection against cucumber mosaic virus (CMV) and pepper mild mottle virus (PMMov) in the local lesion or systemic infection assays. LDH-based nanoparticle technology can also be used in similar way to offer insect protection in plants.
Cost-efficient methods for the production of vast amounts of dsRNA are being optimized and include bacterial, plant, and synthetic production (Palli 2014; Andrade and Hunter 2016). The use of bacteria to synthesize and deliver dsRNA is being pursued for managing agricultural pests especially in non-major crops, such as vegetables and fruits, and for developing insecticidal baits for urban pests, such as ants, cockroaches, and termites (Zhou et al. 2008; Ratzka et al. 2013). The delivery of dsRNA through bacteria and viruses and improving the RNAi efficiency through use of nanoparticles, liposomes, and/or chemical modifications are discussed in Joga et al. (2016). Many of the main crop pest species have already been targeted by RNAi technology using various genes and delivery methods. The first RNAi-based product as a spray is expected for market release during 2017/18 (Joga et al. 2016).
4.1 Coleoptera
RNAi is quite effective in insects that belong to order Coleoptera (Tomoyasu et al. 2008), and this fact has been corroborated through studies conducted in different species from the order. The utility of RNAi in both basic and applied science has been demonstrated in the beetle species, and they appear to be the first target group to be controlled by the new generation of RNAi transgenics (Palli 2012; Palli 2014; Rodrigues and Figueira 2016). Small quantities of ingested dsRNA appears to be sufficient to initiate RNAi response in beetles, such as the western corn rootworm, Colorado potato beetle, southern corn rootworm, Diabrotica undecimpunctata howardi, and the canola flea beetle, Phyllotreta striolata (Baum et al. 2007; Bolognesi et al. 2012; Zhao et al. 2008; Baum and Roberts 2014), which was evident by low LC50 values (1–10 ppb). This level of sensitivity to ingested dsRNA was not observed in insect species outside the order Coleoptera, and the sensitivity was exhibited by both the larval and adult stages (Rangasamy and Siegfried 2012; Zhao et al. 2008), although the in vivo amplification of dsRNA/siRNA has not been shown in beetles. While systemic RNAi is functional in most beetle species studied, variable RNAi responses have been documented across the species, notably to orally delivered dsRNA. RNAi does not seem to work uniformly in all beetles, as illustrated in studies with red flour beetle, T. castaneum, and cotton boll weevil, Anthonomus grandis (Baum et al. 2007; Whyard et al. 2009). The western corn rootworm is one of the most important agricultural pests in which plant-mediated RNAi was successfully demonstrated. Significant larval stunting and mortality were observed in the WCR feeding on maize roots that express a hairpin version of the housekeeping gene vacuolar ATPase (vATPase). The maize roots showed less injury as well (Baum et al. 2007). Comprehensive studies of RNAi in corn rootworm by Baum et al. (2007) provided important insights into the parameters for successful RNAi, for example, their study showed that screening a large number of gene targets through simple surface overlay diet bioassays at relatively low concentration of ~50 ng/cm2 was effective in identifying suitable targets causing lethal phenotypes for successful environmental RNAi in corn rootworm. At least 2/5 of the total 290 gene targets screened were found to cause rootworm mortality or stunting (Baum et al. 2007; Baum et al. 2011). Specifically, the study by Baum et al. (2007) reported no significant difference in efficacy of six ~300 bp dsRNAs corresponding to the V-ATPase region in target gene knockdown in WCR suggesting that a single dsRNA of this size is optimum for RNAi in rootworms. Growth inhibition and mortality were observed in adults fed with vATPase dsRNA-treated artificial diet containing the feeding stimulant cucurbitacin as bait (Rangasamy and Siegfried 2012). In this study, mRNA levels were found to decrease within 24 h of ingestion of dsRNA; however, decrease in protein levels was observed only after 3 days of feeding. The RNAi effect resulted in mortality, although complete suppression of protein was not achieved. A detailed study of the corn rootworm Snf7 ortholog (DvSnf7), which encodes an essential protein involved in intracellular trafficking showed that dsRNAs of greater than or equal to approximately 60 base pairs (bp) are required for the initiation of biological activity in artificial diet bioassays (Bolognesi et al. 2012). Additionally, 21bps short interfering (si) RNAs are not taken up by the midgut cells and therefore failed to trigger the silencing of Snf7 gene, supporting the size versus activity relationship observed in diet bioassays.
Parental RNAi (pRNAi) is an RNA interference response where the gene knockdown phenotype is observed in the progeny of the treated organism (Vélez et al. 2017). In this type of RNAi, the uptake of dsRNA that targets genes regulating embryonic development by adults results in reduced egg hatch rates or complete absence of viable larvae, with the adults remaining unaffected (Khajuria et al. 2015; Fishilevich et al. 2016). A recent study by Vélez et al. (2017) probed the parameters for successful parental RNAi in WCR for two target genes the chromatin remodeling gene brahma (brm) and the gap gene hunchback (hb). The parameters investigated included the concentration, duration, and timing of exposure, with respect to the mating status in WCR females, and the effects of brm and hb dsRNA on male sperm viability and fecundity were also evaluated. Results from this study demonstrate that all parameters studied affect the strength of pRNAi phenotype in females and very subtle effects on sperm count were observed in males. These diet-based pRNAi studies thus provide a framework for developing the technology for field level testing of plant-based pRNAi. Hu et al. (2016) reported the discovery of two new gene targets, the dvssj1 and dvssj2, in WCR that are orthologs of Drosophila genes snakeskin (ssk) and mesh, respectively. Oral delivery of dsRNA targeting dvssj1 and dvssj2 through diet-based insect feeding assays demonstrated target gene suppression, larval growth inhibition, and mortality. Transgenic plants expressing dsRNA of dvssj1 were protected from WCR damage and showed insecticidal activity.
Since the leading insect model organism, D. melanogaster, lacks a robust systemic RNAi response, Tomoyasu et al. (2008) analyzed the genes involved in RNA-mediated gene silencing and the systemic RNAi response in T. castaneum. These studies showed that T. castaneum contains a relatively larger inventory of core component genes than D. melanogaster that probably is responsible for the observed sensitivity of this coleopteran species to dsRNA. Functional analysis of three Tribolium homologues of C. elegans sid-1 genes suggested that T. castaneum sid-like genes are not required for systemic RNAi. Target genes having clear RNAi phenotypes in the model insect T. castaneum were studied further in D. v. virgifera larvae, to test the efficacy of RNAi for target-site screening. Delivery of dsRNA of D. v. virgifera orthologs of laccase 2 (DvvLac2) and chitin synthase 2 (DvvCHS2) by injection resulted in prevention of post-molt cuticular tanning and reduced chitin levels in midguts, respectively, thus providing a tool for identifying potential insecticidal target in western corn rootworm (Alves et al. 2010).
Colorado potato beetle (CPB), a notorious insect pest on solanaceous vegetables potatoes, tomatoes, and eggplants, has not only developed resistance against insecticides but also has an exceptional ability to detoxify plant chemicals. Transgenic potato plants expressing dsRNA targeting CPB genes demonstrated limited success. Feeding dsRNA expressed in bacteria was found to work very well in killing CPB (Palli 2014). In insects species where long dsRNA is more effective than siRNA for effective environmental RNAi, plant-mediated RNAi may not be effective against insect herbivory if there are low levels of dsRNA in the tissue due to the presence of the endogenous plant RNAi pathways that processes dsRNAs into short interfering RNAs. This bottleneck was addressed by expressing the dsRNA in chloroplasts (which lack an RNAi machinery), thereby improving the levels of dsRNA needed to provide protection (Zhang et al. 2015). Transplastomic potato plants expressing hairpin versions of B-actin and Shrub genes in the chloroplasts conferred complete plant protection from insect herbivory; a dramatic increase in the dsRNA levels that induced up to 100% mortality of the CPB in only 5 days was also observed in these plants.
4.2 Lepidoptera
The order Lepidoptera (moths, butterflies, and skippers) represents not only the second largest order in the class Insecta, but also includes major pests of agricultural importance (Xu et al. 2016). The lepidopteran insect herbivores were successfully managed by the first-generation insecticidal plants expressing the Bt proteins for nearly two decades; however, recent reports of resistance evolution to Bt proteins have created the need to find alternatives to manage these pests. Consequently, the lepidopterans were one of the first and main targets for RNAi crops. However, studies have highlighted that in Lepidoptera RNAi has many times proven to be difficult to achieve, and a large variation in response was observed across species (Terenius et al. 2011; Xu et al. 2016). Furthermore, a review of the experimental data of RNAi in Lepidoptera also revealed interesting trends, viz., RNAi is particularly effective in the family Saturniidae, genes involved in immunity are good RNAi targets, knockdown of gene expression in the epidermal tissue seems to be the most difficult to achieve, and high dsRNA dosages are needed for silencing genes by oral delivery of dsRNA (Terenius et al. 2011). Comparative studies of RNAi response in the Coleopteran (robust RNAi response) and Lepidopteran insects (poor RNAi response) undertaken to understand the varying RNAi efficiency in these two insect orders suggest that despite efficient uptake of dsRNA by the Lepidopteran and Coleopteran cell lines, the dsRNA was degraded faster in the Lepidoptera. Furthermore, experimental evidence showed that the dsRNA was processed to siRNA in Coleoptera but not in Lepidoptera thus suggesting that dsRNA degradation, poor intracellular transport of dsRNA, reduced accessibility of dsRNA to the RNAi machinery, and reduced activity of the RNAi machinery are the likely factors for poor RNAi response in Lepidopteran insects (Shukla et al. 2016).
The first lepidopteran RNAi experiments were reported in 2002, where the knockdown of a pigment gene following the dsRNA injection into B. mori embryos (Quan et al. 2002), the silencing of hemolin gene expression by heritable RNAi in H. cecropia embryos (Bettencourt et al. 2002), and a study on a putative Bacillus thuringiensis toxin receptor in Spodoptera litura (Rajagopal et al. 2002) were reported. An increasing number of reports were published since 2007, and particularly after 2010, that describe the successful RNAi experiments in moth species using classic and novel dsRNA delivery methods, viz., microinjection, feeding, soaking, electroporation, and transgenic insect technique, as well as viral-mediated, bacterial-mediated, and plant-mediated dsRNA uptake. A comprehensive review of the recent progresses of the RNAi technique in moths has been published by Xu et al. (2016). Chemically synthesized dsRNA or siRNA was used to demonstrate and validate gene function in initial studies in Lepidoptera, and in later studies it was used to identify suitable targets for RNAi-based crop protection. In all of these studies, the dsRNA was delivered to the insects either by incorporation of the dsRNA into artificial diet, by droplet feeding, or by treating the leaf tissue prior to feeding. The first example of gene silencing via oral delivery of dsRNA was reported by Turner et al. (2006) in the brown apple moth, Epiphyas postvittana (Walker), to achieve suppression of several target genes by droplet feeding of 4000 ppm dsRNA solution. Subsequently, silencing via oral uptake of dsRNA was reported in a wide range of lepidopteran species, but the objective of these studies was not to illustrate the RNAi-mediated insect control or mortality but to investigate gene function in a metabolic or developmental process by selectively suppressing the target gene expression (Belles 2010). Some examples of these early RNAi studies mediated by oral delivery of dsRNA include the down-regulation of a cytochrome P450, CYP6BG1 in the diamondback moth, Plutella xylostella, which reduced the larval resistance to permethrin (Bautista et al. 2009); the knockdown (63–64% reduction of transcript level) of a gut-specific chitinase gene (OnCht) in the European corn borer (ECB), Ostrinia nubilalis larvae, which facilitated an understanding of the regulation of chitin content in the peritrophic matrix (PM) of ECB (Khajuria et al. 2010); the suppression of β1 subunit integrin (βSe1) gene expression from the beet armyworm, S. exigua, to study its role in cellular immune response and larval development (Surakasi et al. 2011); and a 5 μL drop of 3μg SfT6 dsRNA was used in feeding assays to demonstrate the role of a serine protease gene in the processing of the B. thuringiensis Cry1Ca1 insecticidal protein in the fall armyworm (Rodriguez et al. 2010). High concentrations of dsRNA (50–2500 ppm) were used in all cases, and a concentration-dependent mortality was reported upon silencing of the βSe1 subunit gene in the beet armyworm (Surakasi et al. 2011) and the vacuolar ATPase E subunit gene in M. sexta (Whyard et al. 2009). A vacuolar ATPase-A gene and an arginine kinase gene were targeted in the tomato leafminer, Tuta absoluta, an invasive lepidopteran insect pest that is a major threat to commercial tomato production worldwide causing yield losses of up to 100% in various regions (Desneux et al. 2011; Camargo Barbosa et al. 2016). The uptake of dsRNA by the moth larvae from tomato leaflets treated with in vitro synthesized dsRNA resulted in approximately 60% reduction in transcript accumulation in the larvae for both the targets selected, increased larval mortality and protection against insect herbivory (Camargo Barbosa et al. 2016).
Several studies in Lepidoptera also used chemically synthesized siRNA to suppress target gene expression because of the difficulties faced in delivering sufficient dsRNA to the Lepidopteran gut epithelial cells (Gong et al. 2011). Feeding siRNAs specific to acetylcholine esterase AChE to H. armigera larvae at ~0.35 ppm along with the artificial diet resulted in a 15% increase in insect mortality followed by other phenotypes which include growth inhibition of larvae, pupal weight reduction, malformation, and lower fecundity as compared to the control larvae (Kumar et al. 2009). Similar results were reported when acetylcholine esterase genes AChE1 and AChE2 genes were targeted in P. xylostella using chemically synthesized and modified siRNAs (Gong et al. 2013). The siRNAs were modified by addition of a dTdT overhang in the 3′end, 2′-methyl-nucleotides, and 5′ polyethylene glycol (PEG), and this sodium salt formulation contained chitosan. In this study it was found that silencing of PxAChE2 caused higher mortality compared to PxAChE1, thus confirming the importance of PxAChE2 in P. xylostella. In the laboratory cabbage leaf bioassays, one siRNA, Si-ace2_001, at a concentration of 3 μg cm−2 displayed the best insecticidal activity causing 89.0% mortality and exhibited LC50 and LC90 values of 53.7 μg/mL and 759.71 μg/mL, respectively (Gong et al. 2013). In the field evaluation, P. xylostella larvae feeding on Brassica oleracea and B. alboglabra treated with different siRNA doses had no negative effects on plant morphology, color, and growth of vein; however, Si-ace2_001 in the dose of 200 ppm was moderately harmful to the larvae with a mortality of 58.8% 5 days after exposure (Gong et al. 2013). These studies suggest that siRNA can be readily taken up by insect larvae with their diet, and there might not be a strict dsRNA size dependency to the environmental RNAi in lepidopterans (Baum and Roberts 2014).
The uptake of large dsRNA expressed in Escherichia coli has also been reported to impact the growth and survival of the lepidopteran larvae. In S. exigua, silencing a chitin synthase A by feeding the larvae with bacterial culture expressing dsRNA caused larval mortality to increase by 14%, 21%, 26%, and 18% in the first-instar larvae, fourth and fifth larval instars, the prepupae, and pupae, respectively (Tian et al. 2009). Silencing the CYP6B6 gene by feeding larvae with bacteria expressing dsRNA caused a 27% increase in larval mortality in H. armigera (Zhang et al. 2013). Targeting the expression of arginine kinase (AK), an important regulation factor of energy metabolism in invertebrates, by delivering the dsRNA to larvae through diet containing bacteria expressing arginine kinase dsRNA caused larval mortality in H. armigera to increase by 2–11% (Qi et al. 2015). A major limitation in these studies was that neither the concentration of dsRNA in diet nor the effect of dsRNA alone was reported (Baum and Roberts 2014). Yang and Han (2014) reported the evaluation of different dsRNA delivery methods in H. armigera and concluded that continuous ingestion of the bacteria expressing dsRNA was detrimental to insect development and survival than naked dsRNA and the naked dsRNA degraded much faster in the midgut than in hemolymph. Feeding-based RNAi mediated by dsRNA expressed in bacteria or synthesized in vitro, of a molt-regulating transcription factor CiHR3 in Sugarcane stem borer, Chilo infuscatellus Snellen, caused significant abnormalities and weight loss in insects within 7 days of treatment (Zhang et al. 2012). However, silencing a juvenile hormone esterase-related gene via bacterial delivery of dsRNA did not result in a phenotype in the corn stalk borer, Sesamia nonagrioides (Kontogiannatos et al. 2013).
Transgenic plants expressing insect-specific dsRNAs have been considered as a promising strategy for improving pest resistance to insect herbivory in crops, and therefore studies were undertaken in Lepidoptera to demonstrate the suppression of gene expression via plant-mediated dsRNA delivery. Most studies reported so far that used transgenic plants have targeted genes in H. armigera to suppress the development and survival of the moth pest (Xu et al. 2016). Significant suppression of the ecdysone receptor (EcR) gene expression was observed in H. armigera larvae that fed on tobacco plants expressing EcR dsRNA, and it resulted in significantly higher lethality (40%) compared to the gfp control group (10%). Moreover, the growth of the larvae fed on leaves of transgenic tobacco plants expressing HaEcR dsRNA was significantly delayed, their body sizes reduced, and the larvae died with significant molting defects (Zhu et al. 2012). Elevated mortality and developmental aberrations were reported in the larvae of the beet armyworm when fed on the same transgenic tobacco tissue, probably because of the shared sequence similarity of the EcR target sequences in these two species (Zhu et al. 2012).
Xiong et al. (2013) reported larval mortality of 22–30% and >50% mass reduction in H. armigera that fed on transgenic tobacco leaf disks expressing the dsRNA of a molt-regulating transcription factor (HaHR3). Transgenic cotton plants expressing a dsRNA derived from the H. armigera gossypol-inducible cytochrome P450 CYP6AE14 did not cause mortality in H. armigera larvae that fed on the transgenic tissue; however the plants showed increased tolerance to insect herbivory (Mao et al. 2011). An increase in larval stunting was achieved by co-delivering a cysteine proteinase to damage the larval peritrophic matrix that led to higher gossypol accumulation (Mao et al. 2013). This study suggests that plant damage by insect herbivory can be mitigated by targeting detoxification mechanism in the insect midgut, since it appears to not require a systemic RNAi response in the insect (Baum and Roberts 2014). A recent study used transgenic tobacco (Nicotiana tabacum var. Xanthi) and tomato (Solanum lycopersicum Mill cv. Pusa early dwarf) plants expressing H. armigera chitinase dsRNA (Mamta and Rajam 2016) to show that RNAi-induced mortality in H. armigera larvae that fed on transgenic tissue increased by up to 45%. Transient expression of the dsRNA of the vacuolar ATPase-A gene and an arginine kinase gene of T. absoluta, in the tomato plants by infiltration of Agrobacterium cells carrying binary plasmids expressing the target gene hairpin constructs, and uptake of dsRNA by the larvae by feeding on this tissue conferred plant protection against insect feeding damage and reduced target transcript accumulation in the larvae and associated lethality. This study provides evidence that RNAi could be a promising alternative approach for the control of T. absoluta (Camargo Barbosa et al. 2016).
The delivery of dsRNA targeting larval stage-specific transcripts, as a topical application at 50 ppm, was found to cause significant gene silencing and larval mortality at 5 days post-spray in the Asian corn borer, Ostrinia furnacalis (Wang et al. 2011). From this study it was inferred that sprayed dsRNA could have either directly penetrated the body wall and reached the target site via the hemolymph or reached the site of action via the tracheoles to produce RNAi effect. It was also reported that significant RNAi-induced lethality was observed in many of the treatments despite the absence of significant gene silencing at day 3; this raises pertinent questions about the role of a non-RNAi mechanism in the effects observed or the sensitivity of the method used for measuring transcript knockdown (Wang et al. 2011).
4.3 Hemiptera
Hemipterans (whiteflies, aphids, leafhoppers, and plant hoppers) representing major agricultural pests of crops have piercing sucking mouthparts that are inserted into the plant vascular system. These pests inflict direct damage to plants by sucking sap and indirect damage as vectors transmitting plant pathogens particularly plant viruses (Price and Gatehouse 2008). The systemic insecticides predominantly used to control sap-sucking insects pests have contributed to insecticide resistance and high residual activity. Since these pests are recalcitrant to Bt proteins, novel control strategies have to be developed to manage them. The development of an RNAi-based trait provides a good option for controlling hemipteran herbivores; however, the outcome of this strategy relies on the effective delivery of the dsRNA through the vascular tissues (Andrade and Hunter 2016).
RNAi experiments have been successfully confirmed in the hemipteran herbivores encompassing several economically important pests such as the whitefly (Bemisia tabaci), the brown plant hopper (Nilaparvata lugens), the pea aphid (A. pisum), and plant bugs. In hemipterans, as with other insect orders, early studies were aimed at studying gene function and not insect mortality or pest control (Belles 2010; Paim et al. 2013). These experiments revealed that the oral delivery of dsRNA for gene silencing is an attractive alternative to microinjection in Hemiptera because of the relatively small size and fragile nature of the immature stages (Baum and Roberts 2014). Furthermore, for using RNAi trait in crop protection, oral uptake is a preferred route for dsRNA delivery to insect body, although microinjection was the typical mode of delivery in many of the successful experiments. Difficulties have also been reported in achieving optimum RNAi in some Hemiptera, including A. pisum and the tarnished plant bug, Lygus lineolaris (Allen and Walker 2012), due to the limited persistency of the RNAi trigger in the insect body. Across all RNAi experimental data available for Hemiptera to date, it has been observed that there is substantial amount of variability in the dietary concentrations of dsRNA required for knocking down gene expression levels and/or obtaining lethal phenotypic effects, and response ranging from very low to complete knockdown of the transcripts was reported (Baum and Roberts 2014; Christiaens and Smagghe 2014). This variation is seen not only between different species within the order but even between experiments conducted within the same organism. It has been observed that the Hemiptera require a much higher dietary concentrations of dsRNA, viz., at least three orders of magnitude higher than the effective concentrations used with the coleopteran species (Baum and Roberts 2014). These studies also provide valuable insights on the best approaches and targets for using RNAi as a pest control strategy.
Gene silencing following ingestion of dsRNA delivered via artificial diets and transgenic plants has been reported for several hemipteran insects. The v-ATPase subunit E gene knockdown in A. pisum and associated mortality (Whyard et al. 2009), suppression of gene expression by 41–48% in N. lugens (Li et al. 2011), the silencing of aquaporin 1 (ApAQP1) gene leading to elevated hemolymph osmotic pressure in A. pisum (Shakesby et al. 2009) and in the same species, depletion in the expression of gap gene hunchback (Aphb), a key regulator in the antero-posterior patterning causing the expression of a lethal phenotype (Mao and Zeng 2012), silencing of the gene trehalose phosphate synthase (NlTPS) gene in N. lugens causing suppression of transcript expression, disturbed development and lethality in the planthoppers (Chen et al. 2010), and the downregulation of the Ecdysone receptor (SaEcR) and ultraspiracle protein (SaUSP) genes of the grain aphid Sitobion avenae F. that impacted aphid survival and fecundity (Yan et al. 2016) are some examples of gene silencing and/or associated phenotypes following ingestion of dsRNA in Hemiptera. Upadhyay et al. (2011) reported silencing of the ribosomal protein L9 (RPL 9) and vacuolar ATPase subunit A in the whitefly, Bemisia tabaci, upon ingestion of dsRNA/siRNA, with the LC50 values of 11.21 and 3.08 μg/mL, respectively. More than 80% mortality was observed when the target gene expression was knocked down in whiteflies, and the insects showed remarkably higher sensitivity to siRNA. Wuriyanghan et al. (2011) demonstrated the induction of specific RNAi effects in the potato/tomato psyllid (B. cockerelli) by using a modified artificial feeding system containing 15% sucrose, food coloring, and Cy™3-labeled dsRNA. Their study reported that significant RNAi effects were observed when dsRNAs were provided at high concentrations (500 ng/μL or 1000 ng/μL), and this observation was consistent with those reported in other insects such as light-brown apple moth (E. postvittana) and pea aphid (A. pisum) (Turner et al. 2006; Shakesby et al. 2009). In grain aphid, S. avenae, feeding large dsRNA for multiple gene targets (selected after transcriptome profiling) at dietary concentration of 7.5 ng/μL resulted in downregulation of target gene expression, mortality, and developmental stunting of the aphids (Zhang et al. 2013). This study reports achieving lethal RNAi phenotypes at relatively low concentrations of dsRNA (7.5 ppm), which will minimize the risks associated with off-target effects of using high dsRNA dosages and thus facilitate the application of plant-mediated RNAi for developing insect-proof plants. Furthermore, a homologue of Coo2 (a protein effector that promotes host plant colonization in A. pisum (Mutti et al. 2008)) from grain aphid SaCoo2, was found to cause increased mortality in the S. avenae RNAi experiments; however, how the knockdown of this gene impacts aphid feeding behavior on artificial diets is ambiguous (Mutti et al. 2008; Zhang et al. 2013). Feeding high concentration of Inhibitor of apoptosis (IAP) dsRNA (1000 ppm) failed to elicit any detrimental effect in L. lineolaris nymphs (Allen and Walker 2012), although silencing of the same target gene via dsRNA injection resulted in mortality (Walker and Allen 2011). In the corn plant hopper, Peregrinus maidis, two genes encoding subunits of P. maidis V-ATPase (V-ATPase B and V-ATPase D) were chosen as RNAi target genes, and two delivery methods, viz., oral delivery (500 ng/uL) and microinjection, were evaluated to investigate the effectiveness of RNAi (Yao et al. 2013). Both methods of dsRNA delivery resulted in knockdown of target transcripts; however, with microinjection a reduction of 27-fold in the normalized abundance of V-ATPase transcript 2 days post-injection was observed as compared to a two-fold reduction after 6 days of oral ingestion. The injection method was more rapid and effective, and although prolonged suppression of (day 6) the VATPase D transcript resulted in a detectable lethal phenotype, it was observed at a time point where significant mortality was observed in the control insects as well (Yao et al. 2013; Baum and Roberts 2014). This experiment highlights the limitation of the artificial diet experiments for RNAi studies in Hemiptera, where the phenotypes caused by environmental RNAi are slow to manifest and keeping the insects alive on an artificial diet for >7 days can be a major challenge. Consequently, plant-mediated RNAi, viz., the in planta production of dsRNA of essential insect genes, has been a preferred method for the proof-of-concept studies in Hemiptera.
The plant-mediated RNAi effects of three genes expressed in the midgut of N. lugens were studied (Zha et al. 2011), and although target gene expression was suppressed in the hoppers feeding on the GM rice plants, no lethal phenotype was detected. RNAi activity was demonstrated by targeting Myzus persicae genes expressed in gut (Rack1, a receptor of activated kinase) and salivary glands (MpCoo2) (Pitino et al. 2011). Gene expression was knocked down by up to 60% when aphids were fed on N. benthamiana leaf disks transiently producing dsRNA corresponding to these genes and on A. thaliana plants stably producing the dsRNAs. A decrease in the fecundity of M. persicae was observed that was consistent with these genes having essential functions; however, no lethal effects were observed. Injection of Coo2 siRNA into pea aphid adults (A. pisum) resulted in a dramatic depletion of the target salivary gland transcript, and the aphids injected with siCoo2-RNA died well before the control aphids injected with green fluorescent protein (Mutti et al. 2006), suggesting the greater efficiency of microinjection over plant-mediated dsRNA uptake for controlling this species. A similar outcome, viz., reduced fecundity but no mortality, was observed when M. persicae fed on A. thaliana expressing dsRNA of a serine protease gene (Bhatia et al. 2012) and tobacco plants expressing dsRNA targeting the hunchback (hb) gene (Mao and Zeng 2014).
Three target genes (Rak1, MpCoo2, and MpPIntO2) with different functions in aphids were selected to study the persistence and trans-generational effects of plant-mediated RNAi in the green peach aphid (Coleman et al. 2015). This study demonstrated that for the three genes examined RNAi-mediated downregulation and persistence levels in the aphids were not influenced either by the gene sequence or the function; however, a continuous supply of dsRNA was required to maintain the RNAi effect since insects lack genes encoding an RNA-dependent RNA polymerase (RdRP), the enzyme necessary for the siRNA amplification step that leads to persistent RNAi effects (Sijen et al. 2001). The finding that the RNAi effect is transferred to the next generation in aphids revealed by the downregulation of target genes in nymphs born from mothers exposed to dsRNA-producing transgenic plants renders plant-mediated RNAi as a powerful tool for aphid control (Coleman et al. 2015). More recently, it was shown that transgenic tobacco lines expressing long dsRNA precursors of v-ATPaseA provided resistance to whiteflies by delivering sufficient siRNA to knockdown the whitefly v-ATPase gene expression. A significant silencing response leading to whitefly mortality was recorded in whiteflies feeding on transgenic plants (Thakur et al. 2014). A comprehensive review of the plant-mediated RNAi studies reveals that the dsRNA produced by the plants is processed into short siRNA molecules by the plants own RNAi machinery. The presence of long dsRNA in the plant phloem was mentioned in only one report. These data suggest that the sap-sucking insects are mainly taking up siRNA rather than longer dsRNA, although it has been suggested that the RNAi machinery in insects mainly responds to dsRNA (Christiaens and Smagghe 2014).
5 RNAi Risk Assessment and Regulation
As RNAi-based technologies for crop improvement, pest control, and therapeutic applications are developed and products utilizing these technologies are gearing up for market release, assessment of perceived risks will be of prime importance. The potential risks associated with the RNAi-based technologies can be categorized and assessed under molecular characterization, food/feed risk assessment, and environmental risk assessment. Molecular characterization focuses on establishing effects due to off-target gene silencing that may occur due to sufficient sequence homology between small RNAs and mRNAs influencing the function or process (Lundgren and Duan 2013; Ramon et al. 2014). Identifying these off-target genes would facilitate to understand associated risks. Off-target gene silencing may occur both in the RNAi-based GM plant and also in the organisms feeding on the plant. These organisms include target pests and nontarget organisms (NTOs). It should be noted that not all off-target silencing events would lead to significant reduction in gene expression and/or result in detectable phenotypic changes (Casacuberta et al. 2014). Non-availability of genome data of plants/varieties being used for introduction of RNAi-based transformation event and genome data of nontarget organisms limits the use of bioinformatics-based approaches for NTO risk assessment.
The food/feed risk assessment process follows comparative approach to identify intended and unintended changes that may occur in a GM plant. The comparative assessment includes proximate analysis, analysis of, compositional characteristics, toxicity, allergenicity, and nutritional characteristics of the GM plant. This strategy for evaluating potential food/feed risks of RNAi-based GM plants is accepted to be appropriate by various regulatory agencies worldwide (FSANZ 2013; US EPA 2014). The choice of which component and characteristics to be chosen for compositional and agronomic evaluation is determined during the hazard identification step of problem formulation process and is partly based on their ability to predict harm. It should be noted that unintended effects are by nature not expected naturally and so are difficult to test for them directly (Ladics et al. 2015; Schnell et al. 2015; Devos et al. 2015).
The environmental and ecological risk assessment has been discussed by various authors (Auer and Frederick 2009; Lundgren and Duan 2013; Ramon et al. 2014; Vélez et al. 2017; Roberts et al. 2015). This primarily includes the adverse effects on nontarget organisms, environmental fate, and risk of resistance evolution in target pests. The potential adverse effects on nontarget organisms can be studied following tier-based approach (US EPA 2007; Rose 2007; EFSA 2010a, b; ILSI-CERA 2011; Romeis et al. 2011, 2013). The tier-based assessment involves controlled laboratory studies in lower tiers to field-based studies in highest tier. The early-tier studies assess the adverse effects due to direct exposure to the insecticidal protein at concentrations that are several folds higher than the environmental exposure concentrations (US EPA 2007; Raybould 2011; Romeis et al. 2013). The potential for adverse ecological effects of MON 87411 maize, which expresses DvSnf7 RNA, was studied and reported by Bachman et al. (2016). An assessment plan with the routes and levels of exposure and testing representative functional taxa was developed, and the potential for toxicity of DvSnf7 RNA was evaluated. The test nontarget organisms (NTOs) included predators, parasitoids, pollinators, and soil biota besides aquatic and terrestrial vertebrate species. Endpoint observations recorded included survival, growth, development, and reproduction, and results of their study demonstrated no adverse effects with any species tested at, or above, the maximum expected environmental concentration (MEEC). All margins of exposure for NTOs were >tenfold the MEEC. They concluded that exposure to DvSnf7 RNA, both directly and indirectly, is safe for NTOs at the expected field exposure levels.
Surrogate species or model species are used to conduct early-tier studies (Romeis et al. 2008). In general the surrogate species are selected based on exposure pathway, knowledge on activity and mode of action, amenability of test system, and availability of test organism. However, when risk assessment of RNAi products are being considered, the surrogate species should be selected based on phylogenetic relationship to the target organism, as the surrogate would likely be susceptible due to sequence homology/similarity (Romeis et al. 2013). This necessitates evaluating additional or different surrogate species from those tested for Bt crops (Vélez et al. 2017). Additionally, the susceptibility or unresponsiveness of a model organism to the dsRNA in environment may help in selecting correct surrogate species for NTO studies (Roberts et al. 2015). Among arthropod orders, coleopterans are more sensitive to dsRNA (Belles 2010), and lepidopterans require high concentrations of dsRNA to elicit a response as compared to coleopterans (Ivashuta et al. 2015). The observed differences in RNAi efficiency in insects make it even more difficult and complex in choosing surrogate or model organisms in the risk assessment process.
In most of the NTO studies, the measurable endpoint has been mortality, and limited information exists on effects other than mortality (Vélez et al. 2017). Considering that the effects of RNAi are not completely understood, endpoints other than mortality need to be considered through standardized methods (Auer and Frederick 2009; Vélez et al. 2017). Recently, the risks of RNAi-based GE crops on a nontarget soil micro-arthropod, Sinella curviseta, a decomposer, were tested through RNAi dietary toxicity assay, and the endpoint measurements included gene expression profiles, survival, and life history traits (Pan et al. 2016). S. curviseta larvae developed significantly faster under the treatments of dsDVV and dsSC than the vehicle control, and results of this study suggest that the impacts of ingested arthropod-active dsRNAs on this representative soil decomposer are negligible. The selection and use of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess unintended effects of RNAi GM plants was studied and reported by Yang et al. (2016). This study will be a critical step toward the development of an in vivo dietary RNAi toxicity assay for assessing the risks associated with RNAi transgenic plants.
Higher-tier studies in semi-field, greenhouse, or open field conditions are undertaken only when an adverse effect is detected in lower-tier studies. Long-term field assessment of effects of Bt cotton and Bt corn showed minor or negligible risks to nontarget species in these ecosystems (Daly and Buntin 2005; Head et al. 2005; Lawo et al. 2009; Li and Romeis 2010; Naranjo 2005; Torres and Ruberson 2005, 2007). In a recent study reported by Ahmad et al. (2016), potential impact of GM corn MON 87411 (expresses insecticidal dsRNA transcript and Cry3Bb1 protein besides CP4EPSPS protein) on nontarget arthropods (NTAs) was evaluated in the field. They evaluated NTA abundance and damage among GM corn and comparators. Twenty taxa met minimum abundance criteria, out of which nine were considered to be representative of corn ecosystems. They conclude that there is no adverse environmental impact of MON87411 on NTAs compared to conventional corn and demonstrate utility of relevant transportable data for risk assessment in other corn regions. The higher-tier studies may not always find adverse impacts on NTOs due to the complexity of ecosystems and effects thereof.
Lundgren and Duan (2013) identified other reputed risks to NTOs based on the pharmaceutical literature such as immune stimulation and over-saturation of the RNAi machinery. However, as discussed by Bachman et al. (2016), the diets of NTOs consist of plant or animal sources which naturally contain dsRNAs, and there exists a long history of safe consumption of these endogenous dsRNA across eukaryotes. With constant oral exposure to environmental dsRNA endogenously present in natural food sources, unintended effects in nontarget organisms from immune stimulation and RNA machinery saturation are extremely unlikely to result from relatively low exposures to dsRNA resulting from cultivation of MON 87411.
The environmental fate of dsRNA can be addressed in terms of stability and persistence in the environment (Heinemann et al. 2011). Recent studies reported by Dubelman et al. (2014) indicate that the biological activity of Snf7 dsRNA is lost within 2 days after application to different types of soil. Also, up to 90% degradation of the applied dsRNA in soil was observed within 35 h. The other potential route of exposure is through food webs and risks associated can be addressed through experiments with primary consumers and RNAi consumption by the same (Roberts et al. 2015). Resistance evolution in insects to RNAi has not been addressed and documented yet. It is anticipated that insects that carry viruses with RNAi suppressors would be at a selective advantage on RNAi-protected crops, and RNAi-based prophylactics for honey bee colonies would select for viral pathogens with RNAi suppression (Scott et al. 2013). These authors also discuss how genetic variability within and among insect populations, mismatch between dsRNA and target transcript, and single-nucleotide polymorphisms (SNPs) could provide selective advantage for resistance evolution.
Although, the specificity and robustness of RNAi have triggered an immense interest in using RNAi as a tool for creating insect-resistant crops, commercialization is likely to be fraught with challenges. Beyond safety issues, a major impediment includes the lack of a comprehensive federal regulatory framework for estimating the environmental and ecological risks posed by these technologies. Technology evaluation is ongoing, and the development of a standardized risk assessment paradigm is being developed concurrently. However, a number of critical gaps remain including off-target effects, environmental fate, and importantly, the risk of resistance evolution in target pests. Concurrent with limitations such as off-target effects, toxicity, and unsafe delivery methods that have to be overcome before RNAi can be considered for widespread commercial applications in agriculture, it is crucial that a risk assessment paradigm that can proactively anticipate potential nontarget effects be developed for pesticidal RNAs prior to the lifting of deregulation of this technology. Studies reported by Bachman et al. (2016), Ahmad et al. (2016), Pan et al. (2016), and Yang et al. (2016) can potentially form a basis for risk assessment of RNAi-based GM plants or products, within the existing regulatory frameworks.
Conclusions
While significant advances in RNAi methods and applications in agriculture have occurred recently, especially against viruses, the efficacy of the approach against insect pests in the field is yet to be fully established. The discovery of RNAi and the subsequent research on RNAi in insects have demonstrated the profound impact that this technology can have not only in understanding gene regulation in insects but developing pest management solutions for protecting plants from insect herbivory. The applications of RNAi have been studied in several target insect pests belonging to orders such as Coleoptera, Lepidoptera, and Hemiptera. These studies paved the way to a better understanding of using RNAi as a pest management tool, while concurrently highlighting the caveats of using this tool for sustainable management of crop pests. The experimental data generated in the laboratory on several targets shows that with the exception of few studies, the phenotypes observed were mostly sublethal, and field-efficacy data is lacking for many targets and species. It is now evident that the effects of dsRNA; both target and off-target; are species-dependent and target gene-dependent. RNAi provides a mode of action unique among insecticidal agents through the mechanism of gene suppression and therefore can complement the current methods deployed for pest control. Although the current regulatory system allows following the existing methods, further refinement may be required in terms of measurement of target and off-target effects. Therefore, the future course of action for deploying this technology on a commercial scale depends on how these challenges are addressed. Also, the technologies that enable effective and efficient RNAi sprays such as BioClay, based on nanoparticles, should be further explored.
References
Abrieux A, Debernard S, Maria A, Gaertner C, Anton S, Gadenne C, Duportets L (2013) Involvement of the G-protein-coupled dopamine/ecdysteroid receptor DopEcR in the behavioral response to sex pheromone in an insect. PLoS One 8:88. doi:10.1371/journal.pone.0072785
Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD et al (2000) The genome sequence of Drosophila melanogaster. Science 287:2185–2195
Agrawal N, Malhotra P, Bhatnagar RK (2004) siRNA-directed silencing of transgene expressed in cultured insect cells. Biochem Biophys Res Commun 320:428–434
Ahmad A, Negri I, Oliveira W, Brown C, Asiimwe P, Sammons B, Horak M, Jiang C, Carson D (2016) Transportable data from non-target arthropod field studies for the environmental risk assessment of genetically modified maize expressing an insecticidal double-stranded RNA. Transgenic Res 25:1–17
Allen ML, Walker WB (2012) Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J Insect Physiol 58:391–396. doi:10.1016/j.jinsphys.2011.12.014
Alves AP, Lorenzen MD, Beeman RW, Foster JE, Siegfried BD (2010) RNA interference as a method for target-site screening in the western corn rootworm, Diabrotica virgifera virgifera. J Insect Sci 10:162. doi:10.1673/031.010.14122
Andrade CE, Hunter WB (2016) RNA interference – natural gene-based technology for highly specific pest control (HiSPeC). In: Abdurakhmonov IY (ed) RNA interference. InTech, Croatia, pp 391–409
Antonio DSM, Guidugli-Lazzarini KR, Do Nascimento AM, ZLP S, Hartfelder K (2008) RNAi-mediated silencing of vitellogenin gene function turns honeybee (Apis mellifera) workers into extremely precocious foragers. Naturwissenschaften 95:953–961. doi:10.1007/s00114-008-0413-9
Arakane Y, Hogenkamp DG, Zhu YC, Kramer KJ, Specht CA, Beeman RW, Kanost MR, Muthukrishnan S (2004) Characterization of two chitin synthase genes of the red flour beetle, Tribolium castaneum, and alternate exon usage in one of the genes during development. Insect Biochem Mol Biol 34:291–304
Arimatsu Y, Kotani E, Sugimura Y, Furusawa T (2007) Molecular characterization of a cDNA encoding extracellular dsRNase and its expression in the silkworm, Bombyx mori. Insect Biochem Mol Biol 37:176–183
Aronstein K, Saldivar E (2005) Characterization of a honey bee Toll related receptor gene Am18w and its potential involvement in antimicrobial immune defense. Apidologie 36:3–14
Aronstein K, Oppert B, Lorenzen M (2011) RNAi in agriculturally-important arthropods. In: Grabowski PP (ed) RNA processing. In Tech, Shanghai, pp 157–180
Asgari S (2013) MicroRNA functions in insect. Insect Biochem Mol Biol 43:388–397
Auer C, Frederick R (2009) Crop improvement using small RNAs: applications and predictive ecological risk assessments. Trends Biotechnol 27:644–651. doi:10.1016/j.tibtech.2009.08.005
Bachman PM, Huizinga KM, Jensen PD, Mueller G, Tan J, Uffman JP, Levine SL (2016) Ecological risk assessment for DvSnf7 RNA: a plant-incorporated protectant with targeted activity against western corn rootworm. Regul Toxicol Pharmacol 81:77–88. doi:10.1016/j.yrtph.2016.08.001
Barnard A-C, Nijhof AM, Gaspar ARM, Neitz AWH, Jongejan F, Maritz-Olivier C (2012) Expression profiling, gene silencing and transcriptional networking of metzincin metalloproteases in the cattle tick, Rhipicephalus (Boophilus) microplus. Vet Parasitol 186:403–414. doi:10.1016/j.vetpar.2011.11.026
Baum JA, Roberts JK (2014) Progress towards RNAi-mediated insect pest management. In: Dhadialla TS, Gill SS (eds) Insect midgut and insecticidal proteins, Advances in insect physiology, vol 47. Academic, London, pp 249–295
Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O et al (2007) Control of coleopter an insect pests through RNA interference. Nat Biotechnol 25:1322–1326. doi:10.1038/nbt1359
Baum JA, Cajacob CA, Feldmann P, Heck GR, Nooren I, Plaetinck G, Maddelein W, Vaughn T (2011) Methods for control of insect infestation in plants and compositions thereof. US Patent No 7, 943,819
Bautista MAM, Miyata T, Miura K, Tanaka T (2009) RNA interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella, reduces larval resistance to permethrin. Insect Biochem Mol Biol 39:38–46
Beck M, Strand MR (2005) Glc1.8 from Microplitis demolitor bracovirus induces a loss of adhesion and phagocytosis in insect high five and S2 cells. J Virol 79:1861–1870
Belles X (2010) Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu Rev Entomol 55:111–128
Bettencourt R, Terenius O, Faye I (2002) Hemolin gene silencing by ds-RNA injected into Cecropia pupae is lethal to next generation embryos. Insect Mol Biol 11:267–271
Bhatia V, Bhattacharya R, Uniyal PL, Singh R, Niranjan RS (2012) Host generated siRNAs attenuate expression of serine protease gene in Myzus persicae. PLoS One 7:e46343. doi:10.1371/journal.pone.0046343
Blandin S, Moita LF, Köcher T, Wilm M, Kafatos FC, Levashina EA (2002) Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep 3:852–856
Bolognesi R, Ramaseshadri P, Anderson J, Bachman P, Clinton W, Flannagan R et al (2012) Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS One 7:e47534. doi:10.1371/journal.pone.0047534
Borgio JF (2010) RNAi mediated gene knockdown in sucking and chewing insect pests. J Biopesticides 36:153–161
Bronkhorst AW, van Rij RP (2014) The long and short of antiviral defense: small RNA-based immunity in insects. Curr Opin Virol 7:19–28. doi:10.1016/j.coviro.2014.03.010
Camargo Barbosa GO, Possignolo IP et al (2016) RNA interference as a gene silencing tool to control Tuta absoluta in tomato (Solanum lycopersicum). Peer J 4:e2673. doi:10.7717/peerj.2673
Caplen NJ, Fleenor J, Fire A, Morgan RA (2000) dsRNA-mediated gene silencing in cultured Drosophila cells, a tissue culture model for the analysis of RNA interference. Gene 252:95–105
Carrière Y, Crickmore N, Tabashnik BE (2015) Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat Biotechnol 33:161–168
Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136(4):642–655. doi:10.1016/j.cell.2009.01.035
Casacuberta JM, Devos Y, du Jardin P, Ramon M, Vaucheret H, Nogué F (2014) Biotechnological uses of RNAi in plants: risk assessment considerations. Trends Biotechnol 33:145–147. doi:10.1016/j.tibtech.2014.12.003
Chen J, Zhang D, Yao Q, Zhang J, Dong X, Tian H et al (2010) Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Mol Biol 19(6):777–786. doi:10.1111/j.1365-2583.2010.01038.x
Chen X, Tian H, Zou L, Tang B, Hu J, Zhang W (2008) Disruption of Spodoptera exigua larval development by silencing chitin synthase gene A with RNA interference. Bull Entomol Res 98(06):613–619
Choi H, Glatter T, Gstaiger M, Nesvizhskii AI (2012) SAINT-MS1: protein-protein interaction scoring using label-free intensity data in affinity purification-mass spectrometry experiments. J Proteome Res 11:2619–2624
Christiaens O, Smagghe G (2014) The challenge of RNAi-mediated control of hemipterans. Curr Opin Insect Sci 6:15–21. doi:10.1016/j.cois.2014.09.012
Christiaens O, Sweveres L, Smagghe G (2014) DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 53:307–314. doi:10.1016/j.peptides.2013.12.014
Chu CC, Sun W, Spencer JL, Pittendrigh BR, Seufferheld MJ (2014) Differential effects of RNAi treatments on field populations of the western corn rootworm. Pestic Biochem Physiol 110:1–6
Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE (2000) Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A 97:6499–6503
Coleman AD, Wouters RH, Mugford ST, Hogenhout SA (2015) Persistence and transgenerational effect of plant-mediated RNAi in aphids. J Exp Bot 66:541–548
Daly T, Buntin GD (2005) Effects of Bacillus thuringiensis transgenic corn for lepidopteran control on non-target arthropods. Environ Entomol 34:1292–1301. doi:10.1603/0046-225X(2005)034[1292:EOBTTC]2.0.CO;2
Dass CR, Choong PF (2008) Chitosan-mediated orally delivered nucleic acids: a gutful of gene therapy. J Drug Target 16:257–261
Desneux N, Luna MG, Guillemaud T, Urbaneja A (2011) The invasive South American tomato pinworm, Tuta absoluta, continues to spread in Afro-Eurasia and beyond: the new threat to tomato world production. J Pest Sci 84:403–408
Devos Y, Álvarez-Alfageme F, Gennaro A, Mestdagh S (2015) Assessment of unanticipated unintended effects of genetically modified plants on non-target organisms: a controversy worthy of pursuit? J Appl Entomol 140:1–10. doi:10.1111/jen.12248. [Epub ahead of print]
Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson B (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151–156
Dubelman S, Fischer J, Zapata F, Huizinga K, Jiang C, Uffman J et al (2014) Environmental fate of double-stranded RNA in agricultural soils. PLoS One 9:e93155. doi:10.1371/journal.pone.0093155
Dykxhoorn DM, Lieberman J (2005) The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu Rev Med 56:401–423
Dzitoyeva S, Dimitrijevic N, Manev H (2001) Intra-abdominal injection of double-stranded RNA into anesthetized adult Drosophila triggers RNA interference in the central nervous system. Mol Psychiatry 6(6):665–670
EFSA (European Food Safety Authority) (2010a) EFSA panel on plant protection products and their residues (PPR); scientific opinion on the development of specific protection goal options for environmental risk assessment of pesticides, in particular in relation to the revision of the guidance documents on aquatic and terrestrial ecotoxicology (SANCO/3268/2001 and SANCO/10329/2002). EFSA J 8(10):1821. doi:10.2903/j.efsa.2010.1821. (55 pp)
EFSA (European Food Safety Authority) (2010b) EFSA panel on genetically modified organisms (GMO); scientific opinion on the assessment of potential impacts of genetically modified plants on non-target organisms. EFSA J 8(11):1877. doi:10.2903/j.efsa.2010.1877. (72 pp)
Fabrick JA, Kanost MR, Baker JE (2004) RNAi-induced silencing of embryonic tryptophan oxygenase in the Pyralid moth, Plodia interpunctella. J Insect Sci 4(15):9
Farooqui T, Robinson K, Vaessin H, Smith BH (2003) Modulation of early olfactory processing by an octopaminergic reinforcement pathway in the honeybee. J Neurosci 23:5370–5380
Feinberg EH, Hunter CP (2003) Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301:1545–1547
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Fishilevich E, Velez AM, Storer NP, Li HR, Bowling AJ, Rangasamy M, Worden SE, Narva KE, Siegfried BD (2016) RNAi as a management tool for the western corn rootworm, Diabrotica virgifera virgifera. Pest Manag Sci 72:1652–1663. doi:10.1002/ps.4324
FSANZ (2013) Response to Heinemann et al. on the regulation of GM crops and foods developed using gene silencing. http://www.foodstandards.govt.nz/consumer/gmfood/Documents/Heinemann%20Response%20210513.pdf
Garbutt JS, Bellés X, Richards EH, Reynolds SE (2013) Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: evidence from Manduca sexta and Blattella germanica. J Insect Physiol 59:171–178
Gatehouse HS, Gatehouse LN, Malone LA, Hodges S, Tregidga E, Todd J (2004) Amylase activity in honey bee hypopharyngeal glands reduced by RNA interference. J Apic Res 43:9–13
Georghiou GP, Lagunes-Tejeda A (1991) The occurrence of resistance to pesticides in arthropods: an index of cases reported through 1989. Food and Agriculture Organization of the United Nations, Rome
Ghanim M, Kontsedalov S, Czosnek H (2007) Tissue-specific gene silencing by RNA interference in the whitefly, Bemisia tabaci (Gennadius). Insect Biochem Mol Biol 37:732–738. doi:10.1016/j.ibmb.2007.04.006
Ghildiyal M, Zamore PD (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet 10:94–108
Gong L, Chen Y, Hu Z, Hu MY (2013) Testing insecticidal activity of novel chemically synthesized siRNA against Plutella xylostella under laboratory and field conditions. PLoS One 8:88. doi:10.1371/journal.pone.0062990
Gong LA, Yang XQ, Zhang BL, Zhong GH, Hu MY (2011) Silencing of Rieske iron-sulfur protein using chemically synthesised siRNA as a potential biopesticide against Plutella xylostella. Pest Manag Sci 67:514–520. doi:10.1002/ps.2086
Gordon KH, Waterhouse PM (2007) RNAi for insect-proof plants. Nat Biotechnol 25:1231–1232
Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146–1150
Hassanien ITE, Meyering-Vos M, Hoffmann KH (2014) RNA interference reveals allatotropin functioning in larvae and adults of Spodoptera frugiperda (Lepidoptera, Noctuidae). Entomologia 2:56–64. doi:10.4081/entomologia.2014.169
He B, Chu Y, Yin M, Müllen K, An C, Shen J (2013) Fluorescent nanoparticle delivered dsRNA toward genetic control of insect pests. Adv Mater Weinheim 25:4580–4584. doi:10.1002/adma.201301201
Head G, Moar M, Eubanks M, Freeman B, Ruberson J, Hagerty A, Turnipseed S (2005) A multiyear, large-scale comparison of arthropod populations on commercially managed Bt and non-Bt cotton fields. Environ Entomol 34:1257–1266
Heinemann JA, Kurenbach B, Quist D (2011) Molecular profiling – a tool for addressing emerging gaps in the comparative risk assessment of GMOs. Environ Int 37:1285–1293
Horn T, Boutros M (2010) E-RNAi: a web application for the multi-species design of RNAi reagents – 2010 update. Nucleic Acids Res 38:W332–W339
Horn T, Sandmann T, Boutros M (2010) Design and evaluation of genome-wide libraries for RNA interference screens. Genome Biol 11:R61
Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD (2007) The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17:1265–1272
Hu X, Richtman NM, Zhao J-Z, Duncan KE, Niu X, Procyk LA, Oneal MA, Kernodle BM, Steimel JP, Crane VC, Sandahl G, Ritland JL, Howard RJ, Presnail JK, Lu AL, Wu G (2016) Discovery of midgut genes for the RNA interference control of corn rootworm. Sci Rep 6:30542. doi:10.1038/srep30542
Huang JH, Lee HJ (2011) RNA interference unveils functions of the hypertrehalosemic hormone on cyclic fluctuation of hemolymph trehalose and oviposition in the virgin female Blatella germanica. J Insect Physiol 57:858–864
Hunter W, Ellis J, Vanengelsdorp D, Hayes J, Westervelt D, Glick E, Williams M, Sela I, Maori E, Pettis J et al (2010) Large-scale field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae). PLoS Pathog 6:e1001160
Hunter WB, Glick E, Paldi N, Bextine BR (2012) Advances in RNA interference: dsRNA treatment in trees and grapevines for insect pest suppression. Southwest Entomol 37:85–87. doi:10.3958/059.037.0110
Huvenne H, Smagghe G (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol 56(3):227–235. doi:10.1016/j.jinsphys.2009.10.004
ILSI-CERA (2011) Problem formulation for the environmental risk assessment of RNAi plants. International Life Sciences Institute, Center for Environmental Risk Assessment, Washington, D.C.
Ivashuta S, Zhang Y, Wiggins BE, Ramaseshadri P, Segers GC, Johnson S et al (2015) Environmental RNAi in herbivorous insects. RNA 21:840–850. doi:10.1261/rna.048116.114
James C (2014) Global status of commercialized biotech/GM Crops: 2014. ISAAA Brief No. 49. ISAAA, Ithaca, NY
Jaubert-Possamai S, Le T, Bonhomme G, Christophides J, Rispe GK, Tagu D (2007) Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol 7:63
Joga MR, Zotti MJ, Smagghe G, Christiaens O (2016) RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front Physiol 7:553. doi:10.3389/fphys.2016.00553
Jose AM, Hunter CP (2007) Transport of sequence-specific RNA interference information between cells. Annu Rev Genet 41:305–330
Kamath RS, Martinex-Campos M, Zipperlen P, Frasher AG, Ahringer J (2000) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2:research0002
Katoch R, Sethi A, Thakur N, Murdock LL (2013) RNAi for insect control: current perspective and future challenges. Appl Biochem Biotechnol 171(4):847–873
Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, Dostert C, Troxler L, Hetru C, Meignin C, Pfeffer S, Hoffmann JA, Imler JL (2013) Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol 190:650–658
Kennerdell JR, Carthew RW (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017–1026
Khajuria C, Buschman LL, Chen MS, Muthukrishnan S, Zhu KY (2010) A gut-specific chitinase gene essential for regulation of chitin content of peritrophic matrix and growth of Ostrinia nubilalis larvae. Insect Biochem Mol Biol 40:621–629. doi:10.1016/j.ibmb.2010.06.003
Khajuria C, Li H, Narva K, Rangasamy M, Siegfried B (2013) Effectiveness of dsRNA versus siRNA in RNAi mediated gene knock-down in western corn rootworm (Diabrotica virgifera virgifera). In: Program and abstracts, 46th annual meeting of the Society for Invertebrate Pathology Conference on Invertebrate Pathology and Microbial Control. Society for Invertebrate Pathology. Pittsburgh, PA
Khajuria C, Vélez AM, Rangasamy M, Wang H, Fishilevich E, Frey MLF, Carneiro N, Premchand G, Narva KE, Siegfried BD (2015) Parental RNA interference of genes involved in embryonic development of the western corn rootworm, Diabrotica virgifera virgifera LeConte. Insect Biochem Mol Biol 63:54–62
Kitzmann P, Schwirz J, Schmitt-Engel C, Bucher G (2013) RNAi phenotypes are influenced by the genetic background of the injected strain. BMC Genomics 14(1):5. doi:10.1186/1471-2164-14-5
Koch A, Kogel KH (2014) New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol J 12:821–831. doi:10.1111/pbi.12226
Kolliopoulou A, Swevers L (2014) Science direct recent progress in RNAi research in lepidoptera?: intracellular machinery, antiviral immune response and prospects for insect pest control. Curr Opin Insect Sci:1–7. doi:10.1016/j.cois.2014.09.019
Kontogiannatos D, Swevers L, Maenaka K, Park EY, Iatrou K, Kourti A (2013) A functional characterization of a Juvenile Hormone esterase related gene in the moth Sesamia nonagrioides through RNA interference. PLoS One 8:88. doi:10.1371/journal.pone.0073834
Kumar M, Gupta GP, Rajam MV (2009) Silencing of acetylcholinesterase gene of Helicoverpa armigera by siRNA affects larval growth and its life cycle. J Insect Physiol 55:273–278. doi:10.1016/j.jinsphys.2008.12.005
Ladics GS, Bartholomaeus A, Bregitzer P, Doerrer NG, Gray A, Holzhauser T et al (2015) Genetic basis and detection of unintended effects in genetically modified crop plants. Transgenic Res 24:587–603. doi:10.1007/s11248-015-9867-7
Lawo NC, Wackers FL, Romeis J (2009) Indian Bt cotton varieties do not affect the performance of cotton aphids. PLoS One 4:e4804
Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW (2004) Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117:69–81
Li X, Zhang M, Zhang H (2011) RNA interference of four genes in adult Bactrocera dorsalis by feeding their dsRNAs. PLoS One 6(3):e17788. doi:10.1371/journal.pone
Li Y, Romeis J (2010) Bt maize expressing Cry3Bb1 does not harm the spider mite, Tetranychus urticae, or its ladybird beetle predator, Stethorus punctillum. Biol Control 53:337–344. doi:10.1016/j.biocontrol.2009.12.003
Liu F, Wang X, Zhao Y, Li Y, Liu Y, Sun J (2015) Silencing the HaAK gene by transgenic plant-mediated RNAi impairs larval growth of Helicoverpa armigera. Int J Biol Sci 11:67–74. doi:10.7150/ijbs.10468
Liu S, Ding Z, Zhang C, Yang B, Liu Z (2010) Gene knockdown by introthoracic injection of double-stranded RNA in the brown planthopper, Nilaparvata lugens. Insect Biochem Mol Biol 40:666–671
Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q (2009) C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 325:750–753
Loy DL, Mogler MA, Loy DS, Janke B, Kamrud K, Scura ED, Harris DLH, Bartholomay LC (2012) Double-stranded RNA provides sequence dependent protection against infectious myonecrosis virus in Litopenaeus vannamei. J Gen Virol 93:880–888
Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA (2003) Science 299:2039–2045
Lundgren JG, Duan JJ (2013) RNAi-based insecticidal crops. Bioscience 63:657–665. doi:10.1525/bio.2013.63.8.8
Luo Y, Wang X, Yu D, Kang L (2012) The SID-1 double-stranded RNA transporter is not required for systemic RNAi in the migratory locust. RNA Biol 9:663–671
Mamta RKR, Rajam MV (2016) Targeting chitinase gene of Helicoverpa armigera by host-induced RNA interference confers insect resistance in tobacco and tomato. Plant Mol Biol 90:281–292. doi:10.1007/s11103-015-0414-y
Mao J, Zeng F (2012) Feeding-based RNA interference of a gap gene is lethal to the pea aphid, Acyrthosiphon pisum. PLoS One 7:e48718
Mao J, Zeng F (2014) Plant-mediated RNAi of a gap gene-enhanced tobacco tolerance against the Myzus persicae. Transgenic Res 23:145–152. doi:10.1007/s11248-013-9739-y
Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25(11):1307–1313
Mao YB, Tao XY, Xue XY, Wang LJ, Chen XY (2011) Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Res 20:665–673. doi:10.1007/s11248-010-9450-1
Mao YB, Xue XY, Tao XY, Yang CQ, Wang LJ, Chen XY (2013) Cysteine protease enhances plant-mediated bollworm RNA interference. Plant Mol Biol 83:119–129. doi:10.1007/s11103-013-0030-7
Martin D, Maestro O, Cruz J, Mane-Padros D, Belles X (2006) RNAi studies reveal a conserved role for RXR in molting in the cockroach Blattella germanica. J Insect Physiol 52:410–416
Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431(7006):343–349
Miguel SK, Scott JG (2016) The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manag Sci 72(4):801–809. doi:10.1002/ps.4056
Miller SC, Brown SJ, Tomoyasu Y (2008) Larval RNAi in Drosophila? Dev Genes Evol 218:505–510
Mitter N, Elizabeth AW, Karl ER, Li P, Jain RG et al (2017) Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nature Plants 3:16207. doi:10.1038/nplants.2016.207
Mohammed A, Diab MR, Abd-Alla SMM, Hussien EHA (2015) RNA interference – mediated knockdown of vacuolar ATPase genes in pink bollworm, Pectinophora gossypiella. Int J Biol Pharm Allied Sci 4:2641–2660
Mohr S, Perrimon N (2012) RNAi screening: new approaches, understandings and organisms. Wiley Interdiscip Rev RNA 2:145–158
Mutti NS, Park Y, Reese JC, Reeck GR (2006) RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. J Insect Sci 6:1–7. doi:10.1673/031.006.3801
Mutti NS, Louis J, Pappan LK, Pappan K, Begum K, Chen MS, Park Y, Dittmer N, Marshall J, Reese JC, Reeck GR (2008) A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc Natl Acad Sci U S A 105:9965–9969
Mysore K, Andrews E, Li P, Duman-Scheel M (2014) Chitosan/siRNA nanoparticle targeting demonstrates a requirement for single-minded during larval and pupal olfactory system development of the vector mosquito Aedes aegypti. BMC Dev Biol 14:1. doi:10.1186/1471-213X-14-9
Naito Y, Yamuda T, Mastumiya T, Kumiko UT, Saigo K, Morishita S (2005) dsCheck: highly sensitiveoff-target search software for double-stranded RNA-mediated RNA interference. Nucleic Acids Res 33:W589–W591
Nakamura T, Mito T, Miyawaki K, Ohuchi H, Noji S (2008) EGFR signaling is required for re-establishing the proximodistal axis during distal leg regeneration in the cricket Gryllus bimaculatus nymph. Dev Biol 319:46–55
Naranjo SE (2005) Long-term assessment of the effects of transgenic Bt cotton on the abundance of nontarget arthropod natural enemies. Environ Entomol 34:1193–1210
Nayak A, Tassetto M, Kunitomi M, Andino R (2013) RNA interference-mediated intrinsic antiviral immunity in invertebrates. Curr Top Microbiol Immunol 371:183–200
Osanai-Futahashi M, Tatematsu K-I, Futahashi R, Narukawa J, Takasu Y, Kayukawa T, Shinoda T, Ishige T, Yajima S, Tamura T, Yamamoto K, Sezutsu H (2016) Positional cloning of a Bombyx pink-eyed white egg locus reveals the major role of cardinal in ommochrome synthesis. Heredity 116:135–145. doi:10.1038/hdy.2015.74
Paim RM, Araujo RN, Lehane MJ, Gontijo NF, Pereira MH (2013) Long-term effects and parental RNAi in the blood feeder, Rhodnius prolixus (Hemiptera; Reduviidae). Insect Biochem Mol Biol 43:1015–1020. doi:10.1016/j.ibmb.2013.08.008
Palli SR (2012) RNAi methods for management of insects and their pathogens. CAB Rev 7:1–10
Palli SR (2014) RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Curr Opin Insect Sci 6:1–8. doi:10.1016/j.cois.2014.09.011
Pan H, Xu L, Noland JE, Li H, Siegfried BD, Zhou X (2016) Assessment of potential risks of dietary RNAi to a soil micro-arthropod, Sinella curviseta Brook (Collembola: Entomobryidae). Front Plant Sci 7:1028. doi:10.3389/fpls.2016.01028
Perrimon N, Mathey-Prevot B (2007) Applications of high-throughput RNA interference screens to problems in cell and developmental biology. Genetics 175:7–16
Peters L, Meister G (2007) Argonaute proteins: mediators of RNA silencing. Mol Cell 26(5):611–623
Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA (2011) Silencing of aphid genes by dsRNA feeding from plants. PLoS One 6:e25709
Pradhan SK, Nayak DK, Mohanty S, Behera L, Barik SR, Pandit E, Lenka S, Anandan A (2015) Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 8:19. doi:10.1186/s12284-015-0051-8
Price DRG, Gatehouse JA (2008) RNAi-mediated crop protection against insects. Trends Biotechnol 26(7):393–400
Pridgeon JW, Zhao L, Becnel JJ, Strickman DA, Clark GG, Linthicum KJ (2008) Topically applied AaeIAP1 double-stranded RNA kills female adults of Aedes aegypti. J Med Entomol 45:414–420
Qi XL, Su XF, Lu GQ, Liu CX, Liang GM, Cheng HM (2015) The effect of silencing arginine kinase by RNAi on the larval development of Helicoverpa armigera. Bull Entomol Res 105:555–565. doi:10.1017/S0007485315000450
Qian D, Tian L, Qu L (2015) Proteomic analysis of endoplasmic reticulum stress responses in rice seeds. Sci Rep 5:14255. doi:10.1038/srep14255
Qiu S, Adema CM, Lane T (2005) A computational study of off-target effects of RNA interference. Nucleic Acids Res 33:1834–1847
Quan GX, Kanda T, Tamura T (2002) Induction of the white egg 3 mutant phenotype by injection of the double-stranded RNA of the silkworm white gene. Insect Mol Biol 11:217–222
Rajagopal S, Sivakumar N, Agrawal P, Malhotra P, Bhatnagar RK (2002) Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J Biol Chem 277:46849–46851
Ramon M, Devos Y, Lanzoni A, Liu Y, Gomes A, Gennaro A et al (2014) RNAi-based GM plants: food for thought for risk assessors. Plant Biotechnol J 12:1271–1273. doi:10.1111/pbi.12305
Rangasamy M, Siegfried BD (2012) Validation of RNA interference in western corn rootworm Diabrotica virgifera virgifera LeConte (Coleoptera, Chrysomelidae) adults. Pest Manag Sci 68:587–591. doi:10.1002/ps.2301
Ratzka C, Gross R, Feldhaa H (2013) Systemic gene knockdown in Camponotus floridanus workers by feeding of dsRNA. Insect Soc 60(4):475–484. doi:10.1007/s00040-013-0314-6
Raybould A (2011) The bucket and the searchlight: formulating and testing risk hypotheses about the weediness and invasiveness potential of transgenic crops. Environ Biosaf Res 9:123–133
Rinkevich FD, Scott JG (2013) Limitations of RNAi of a6 nicotinic acetylcholine receptor subunits for assessing the in vivo sensitivity to spinosad. Insect Sci 20:101–108
Roberts AF, Devos Y, Lemgo GNY, Zhou X (2015) Biosafety research for non-target organism risk assessment of RNAi-based GE plants. Front Plant Sci 6:958. doi:10.3389/fpls.2015.00958
Rodrigues TB, Figueira A (2016) Management of insect pest by RNAi – a new tool for crop protection. In: Abdurakhmonov IY (ed) RNA interference. InTech, Croatia. doi:10.5772/61807
Rodriguez CL, Trujillo BD, Borras HO, Wright DJ, Ayra-Pardo C (2010) RNAi-mediated knockdown of a Spodoptera frugiperda trypsin-like serine-protease gene reduces susceptibility to a Bacillus thuringiensis Cry1Ca1 protoxin. Environ Microbiol 12:2894–2903. doi:10.1111/j.1462-2920.2010.02259.x
Roignant JY, Carré C, Mugat B, Szymczak D, Lepesant JA, Antoniewski C (2003) Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9:299–230
Romeis J, Bartsch D, Bigler F, Candolfi MP, Gielkens MMC et al (2008) Assessment of risk of insect-resistant transgenic crops to nontarget arthropods. Nat Biotechnol 26(2):203–208. doi:10.1038/nbt1381
Romeis J, Hellmich RL, Candolfi MP, Carstens K, De Schrijver A et al (2011) Recommendations for the design of laboratory studies on non-target arthropods for risk assessment of genetically engineered plants. Transgenic Res 20:1–22. doi:10.1007/s11248-010-9446-x
Romeis J, Raybould A, Bigler F, Candolfi MP, Hellmich RL, Huesing JE, Shelton AM (2013) Deriving criteria to select arthropod species for laboratory tests to assess the ecological risks from cultivating arthropod-resistant genetically engineered crops. Chemosphere 90(2013):901–909
Rose RI (ed) (2007) White paper on tier-based testing for the effects of proteinaceous insecticidal plant-incorporated protectants on non-target invertebrates for regulatory risk assessment. USDA-APHIS and US Environmental Protection Agency, Washington, DC. http://www.epa.gov/pesticides/biopesticides/pips/non-target-arthropods.pdf
Sadeghi A, van Damme EJM, Smagghe G (2009) Evaluation of the susceptibility of the pea aphid, Acyrthosiphon pisum, to a selection of novel biorational insecticides via artificial diet. J Insect Sci 9:65. http://insectscience.org/9.65
Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC (2007) Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev 21:1603–1608
Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH et al (2006) The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol 8:793–802. doi:10.1038/ncbl439
Sarkies P, Miska EA (2013) Is there social RNA? Science 341(6145):467–468. doi:10.1126/science.1243175
Schluns H, Crozier RH (2007) Relish regulates expression of antimicrobial peptide genes in the honeybee, Apis mellifera, shown by RNA interference. Insect Mol Biol 16:753–759
Schnell J, Steele M, Bean J, Neuspiel M, Girard C, Dormann N et al (2015) A comparative analysis of insertional effects in genetically engineered plants: considerations for pre-market assessments. Transgenic Res 24:1–17. doi:10.1007/s11248-014-9843-7
Scott JG, Michel K, Bartholomay LC, Siegfried BD, Hunter WB, Smagghe G, Zhu KY, Douglas AE (2013) Towards the elements of successful insect RNAi. J Insect Physiol 59:1212–1221. doi:10.1016/j.jinsphys.2013.08.014
Shah C, Förstemann K (2008) Monitoring miRNA mediated silencing in Drosophila melanogaster S2-cells. Biochim Biophys Acta 1779:766–772
Shakesby A, Wallace I, Isaacs H, Pritchard J, Roberts D, Douglas A (2009) A water-specific aquaporin involved in aphid osmoregulation. Insect Biochem Mol Biol 39:1–10. doi:10.1016/j.ibmb.2008.08.008
Shukla JN, Kalsi M, Sethi A, Narva KE, Fishilevich E, Singh S et al (2016) Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol 13(7):656–669. doi:10.1080/15476286.2016.1191728
Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107:465–476
Singh AD, Wong S, Ryan CP, Whyard S (2013) Oral delivery of double stranded RNA in larvae of the yellow fever mosquito, Aedes aegypti: implications for pest mosquito control. J Insect Sci 13:1–18
Siomi H, Siomi MC (2009) On the road to reading the RNA-interference code. Nature 457:396–404
Sivakumar S, Rajagopal R, Venkatesh GR, Srivastava A, Bhatnagar RK (2007) Knockdown of aminopeptidase-N from Helicoverpa armigera larvae and in transfected Sf21 cells by RNA interference reveals its functional interaction with Bacillus thuringiensis insecticidal protein Cry1Ac. J Biol Chem 282:7312–7319
Smith P (2013) Delivering food security without increasing pressure on land. Glob Food Sec 2(1):18–23
Soares CAG, Lima CMR, Dolan MC, Piesman J, Beard CB, Zeidner NS (2005) Capillary feeding of specific dsRNA induces silencing of the isac gene in nymphal Ixodes scapularis ticks. Insect Mol Biol 14:443–452
Storer NP, Babcock JM, Schlenz M, Meade T, Thompson GD, Bing JW, Huckaba RM (2010) Discovery and characterization of field resistance to Bt maize: Spodoptera frugiperda (Lepidoptera: Noctuidae) in Puerto Rico. J Econ Entomol 103(4):1031–1038
Sugimoto A (2004) High-throughput RNAi in Caenorhabditis elegans, genome-wide screens and functional genomics. Differentiation 72:81–91
Sun K, Wolters AMA, Loonen AEHM, Huibers RP, van der Vlugt R, Goverse A, Jacobsen E, Visser RGF, Bai Y (2016) Down-regulation of Arabidopsis DND1 orthologs in potato and tomato leads to broad-spectrum resistance to late blight and powdery mildew. Transgenic Res 25:123–138. doi:10.1007/s11248-015-9921-5
Surakasi VP, Mohamed AAM, Kim Y (2011) RNA interference of beta 1 integrin subunit impairs development and immune responses of the beet armyworm, Spodoptera exigua. J Insect Physiol 57:1537–1544
Suzuki Y, Truman JW, Riddiford LM (2008) The role of Broad in the development of Tribolium castaneum: implications for the evolution of the holometabolous insect pupa. Development 135:569–577. doi:10.1242/dev.015263
Swevers L, Smagghe G (2012 ) Use of RNAi for control of insect crop pests. In: Smagghe G, Diaz I, editors. Arthropod-plant interactions: novel insights and approaches for IPM. Dordrecht: Springer; pp. 177–197.
Swevers L, Broeck JV, Smagghe G (2013) The possible impact of persistent virus infection on the function of the RNAi machinery in insects: a hypothesis. Front Physiol 4:319. doi:10.3389/fphys.2013.00319
Szittya G, Burgyan J (2013) RNA interference-mediated intrinsic antiviral immunity in plants. Curr Top Microbiol Immunol 371:153–181
Tabara H, Grishok A, Mello CC (1998) RNAi in C. elegans: soaking in the genome sequence. Science 282:430–431
Tabashnik BE, Thierry B, Yves C (2013) Insect resistance to Bt crops: lessons from the first billion acres. Nat Biotechnol 31:510–521
Tan EL, Tan TM, Chow VTK, Poh CL (2008) Inhibition of enterovirus 71 in virus-infected mice by RNA interference. Mol Ther 15:1931–1938
Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, Kanginakudru S et al (2011) RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol 57(2):231–245. doi:10.1016/j.jinsphys.2010.11.006
Thakur N, Upadhyay SK, Verma PC, Chandrashekar K, Tuli R, Singh PK (2014) Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase a gene. PLoS One 9:88. doi:10.1371/journal.pone.0087235
Tian H, Peng H, Yao Q, Chen H, Xie Q, Tang B et al (2009) Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS One 4(7):e6225. doi:10.1371/journal.pone.0006225
Timmon L, Fire A (1998) Specific interference by ingested dsRNA. Nature 395:854
Timmons L, Court DL, Fire A (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263:103–112
Tomizawa M, Noda H (2013) High mortality caused by high dose of dsRNA in the green rice leafhopper, Nephotettix cincticeps (Hemiptera: Cicadellidae). Appl Entomol Zool 48:553–559
Tomoyasu Y, Denell RE (2004) Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev Genes Evol 214:575–578
Tomoyasu Y, Miller SC, Tomita S, Schoppmeier M, Grossmann D et al (2008) Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol 9:R10. doi:10.1186/gb-2008-9-1-r10
Torres JB, Ruberson JR (2005) Canopy- and ground- dwelling predatory arthropods in commercial Bt and non-Bt cotton fields: patterns and mechanisms. Environ Entomol 34:1242–1256
Torres JB, Ruberson JR (2007) Abundance and diversity of ground-dwelling arthropods of pest management importance in commercial Bt and non-Bt cotton fields. Ann Appl Biol 150:27–39
Turner CT, Davy MW, MacDiarmid RM, Plummer KM, Birch NP, Newcomb RD (2006) RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol Biol 15:383–391
Ulrich J, Dao VA, Majumdar U, Schmitt-Engel C, Schwirz J, Schultheis D et al (2015) Large scale RNAi screen in Tribolium reveals novel target genes for pest control and the proteasome as prime target. BMC Genomics 16:674. doi:10.1186/s12864-015-1880-y
Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C et al (2006) Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem 281:14370–14375. doi:10.1074/jbc.M513868200
Upadhyay SK, Chandrashekar K, Thakur N, Verma PC, Borgio JF, Singh PK et al (2011) RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J Biosci 36:153–161. doi:10.1007/s12038-011-9009-1
US EPA (2007) White paper on tier-based testing for the effects of proteinaceous insecticidal plant-incorporated protectants on non-target arthropods for regulatory risk assessments. United States Environmental Protection Agency, Washington, DC
US EPA (2014) Scientific advisory panel minutes No. 2014-02 (Arlington, VA), 1–77. Available online at: http://www.epa.gov/scipoly/sap/meetings/2014/january/012814minutes.pdf
Valdes VJ, Sampieri A, Sepulveda J, Vaca L (2003) Using double-stranded RNA to prevent in vitro and in vivo viral infections by recombinant baculovirus. J Biol Chem 278:19317–19324
Valdes VJ, Athie A, Salinas LS, Navarro RE, Vaca L (2012) Correction: CUP-1 is a novel protein involved in dietary cholesterol uptake in Caenorhabditis elegans. PLoS One 7(8). doi:10.1371/annotation/5a203055-6c15-43b0-96ad-0fbd5eb9b810
Vélez AM, Fishilevich E, Matz N, Storer NP, Narva KE, Siegfried BD (2017) Parameters for successful parental RNAi as an insect pest management tool in western corn rootworm, Diabrotica virgifera virgifera. Genes 8:7. doi:10.3390/genes8010007
Vijayendran D, Airs PM, Dolezal K, Bonning BC (2013) Arthropod viruses and small RNAs. J Invertebr Pathol 114(2):186–195
Walker WB, Allen ML (2011) RNA interference-mediated knockdown of IAP in Lygus lineolaris induces mortality in adult and preadult life stages. Entomol Exp Appl 138:83–92. doi:10.1111/j.1570-7458.2010.01078.x
Wang Y, Zhang H, Li H, Miao X (2011) Second-generation sequencing supply an effective way to screen RNAi targets in large scale for potential application in pest insect control. PLoS One 6:e18644
Whangbo JS, Hunter CP (2008) Environmental RNA interference. Trends Genet 24(6):297–305. doi:10.1016/j.tig.2008.03.007
Whitten MMA, Facey PD, Del Sol R, Fernández-Martínez LT, Evans MC, Mitchell JJ et al (2016) Symbiont-mediated RNA interference in insects. Proc Biol Sci 283:20160042. doi:10.1098/rspb.2016.0042
Whyard S, Singh AD, Wong S (2009) Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem Mol Biol 39:824–832. doi:10.1016/j.ibmb.2009.09.007
Winston WM, Molodowitch C, Hunter CP (2002) Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295:2456–2459. doi:10.1126/science.1068836
Wu F, Wang PY, Zhao QL, Kang LQ, Xia DG, Qiu ZY, Tang SM, Li MW, Shen XJ, Zhang GZ (2016) Mutation of a cuticle protein gene, BmCPG10, is responsible for silkworm non-moulting in the 2nd instar mutant. PLoS One 11:88. doi:10.1371/journal.pone.0153549
Wuriyanghan H, Rosa C, Falk BW (2011) Oral delivery of double-stranded RNAs and siRNAs induces RNAi effects in the potato/tomato psyllid, Bactericerca cockerelli. PLoS One 6:e27736
Xiong Y, Zeng H, Zhang Y, Xu D, Qiu D (2013) Silencing the HaHR3 Gene by transgenic plant-mediated RNAi to disrupt Helicoverpa armigera development. Int J Biol Sci 9:370–381. doi:10.7150/ijbs.5929
Xu J, Wang X-F, Chen P, Liu FT, Zheng SC, Ye H, Mo MH (2016) RNA interference in moths: mechanisms, applications and progress. Genes 7:88. doi:10.3390/genes7100088
Xu W, Han Z (2008) Cloning and phylogenetic analysis of sid-1-like genes from aphids. J Insect Sci 8:30. doi:10.1673/031.008.3001
Xue XY, Mao YB, Tao XY, Huang YP, Chen XY (2012) New approaches to agricultural insect pest control based on RNA interference. Adv Insect Physiol 42:73–117
Yan T, Chen H, Sun Y, Yu X, Xia L (2016) RNA interference of the ecdysone receptor genes EcR and USP in grain aphid (Sitobion avenae F.) affects its survival and fecundity upon feeding on wheat plants. Davies TGE, ed. Int J Mol Sci 17(12):2098. doi:10.3390/ijms17122098
Yang C, Preisse EL, Zhang H, Liu Y, Dai L, Pan H, Pan H, Zhou X (2016) Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front Plant Sci 7:1672. doi:10.3389/fpls.2016.01672
Yang J, Han ZJ (2014) Efficiency of different methods for dsRNA delivery in cotton bollworm (Helicoverpa armigera). J Integr Agric 13:115–123. doi:10.1016/S2095-3119(13)60511-0
Yao J, Rotenberg D, Afsharifar A, Barandoc AK, Whitfield AE (2013) Development of RNAi methods for Peregrinus maidis, the corn planthopper. PLoS One 8:e70243
Yu N, Christiaens O, Liu JM, Niu J, Cappelle K, Caccia S, Huvenne H, Smagghe G (2013) Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci 20:4–14
YuQ LT, Feng G, Yang K, Pang Y (2008) Functional analysis of the putative antiapoptotic genes, p49 and iap4, of Spodoptera litura nucleopolyhedrovirus with RNAi. J Gen Virol 89:1873–1880
Zamore PD (2001) RNA interference: listening to the sound of silence. Nat Struct Mol Biol 8(9):746–750
Zeynep A, Horn T, Boutros M (2005) E-RNAi: a web application to design optimized RNAi constructs. Nucleic Acids Res 33:W582–W588. doi:10.1093/nar/gki468. Web Server issue
Zha W, Peng X, Chen R, Du B, Zhu L, He G (2011) Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS One 6(5):e20504. doi:10.1371/journal.pone.0020504
Zhang H, Li FL, Cheng C, Jiao DX, Zhou Z, Cheng LG (2013) The identification and characterisation of a new deltamethrin resistance-associated gene, UBL40, in the diamondback moth, Plutella xylostella (L). Gene 530:51–56. doi:10.1016/j.gene.2013.07.075
Zhang X, Zhang J, Zhu KY (2010) Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol Biol 19:683–693
Zhang X, Mysore K, Flannery E, Michel K, Severson DW, Zhu KY et al (2015) Chitosan/interfering RNA nanoparticle mediated gene silencing in disease vector mosquito larvae. J Vis Exp 97:52523. doi:10.3791/52523
Zhang Y, Wiggins B, Lawrence C, Petrick J, Ivashuta S, Heck G (2012) Analysis of plant derived miRNAs in animal small RNA datasets. BMC Genomics 13:381
Zhao HM, Yi X, Hu Z, Chen SH, Dong XL, Gong L (2013) RNAi-mediated knockdown of catalase causes cell cycle ar rest in SL-1 cells and results in low survival rate of Spodoptera litura (Fabricius). PLoS One 8:e59527
Zhao YY, Yang G, Wang-PG YMS (2008) Phyllotreta striolata (Coleoptera: Chrysomelidae): arginine kinase cloning and RNAi-based pest control. Eur J Entomol 105:815–822
Zhou XG, Wheeler MM, Oi FM, Scharf ME (2008) RNA interference in the termite Reticulitermes flavipes through ingestion of double-stranded RNA. Insect Biochem Mol Biol 38:805–815. doi:10.1016/j.ibmb.2008.05.005
Zhu F, Xu J, Palli R, Ferguson J, Palli SR (2011) Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag Sci 67:175–182. doi:10.1002/ps.2048
Zhu F, Cui Y, Walsh DB, Lavine LC (2014) Application of RNA interference toward insecticide resistance management. In: Chandrasekar R, Tyagi BK, Gui ZZ, Reeck GR (eds) Short views on insect biochemistry and molecular biology, vol 2. International Book Mission, Academic, Manhattan, pp 595–619. Chapter-27
Zhu J-Q, Liu S, Ma Y, Zhang J-Q, Qi H-S, Wei Z-J et al (2012) Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PLoS One 7:e38572. doi:10.1371/journal.pone.0038572
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Alamalakala, L., Parimi, S., Patel, N., Char, B. (2018). Insect RNAi: Integrating a New Tool in the Crop Protection Toolkit. In: Kumar, D., Gong, C. (eds) Trends in Insect Molecular Biology and Biotechnology. Springer, Cham. https://doi.org/10.1007/978-3-319-61343-7_10
Download citation
DOI: https://doi.org/10.1007/978-3-319-61343-7_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-61342-0
Online ISBN: 978-3-319-61343-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)