Abstract
Finding relevant concepts from a corpus of ontologies is useful in many scenarios, such as document classification, web page annotation, and automatic ontology population. Many millions of concepts are contained in a large number of ontologies across diverse domains. A SPARQL-based query demands the knowledge of the structure of ontologies and the query language, whereas user-friendlier and, simpler keyword-based approaches suffer from false positives. This is because concept descriptions in ontologies may be ambiguous and may overlap. In this paper, we propose a keyword-based concept search framework, which (1) exploits the structure and semantics in ontologies, by constructing contexts for each concept; (2) generates the interpretations of a query; and (3) balances the relevance and diversity of search results. A comprehensive evaluation against the domain-specific BioPortal and the general-purpose Falcons on widely-used performance metrics demonstrates that our system outperforms both.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The current breed of Semantic Web search engines can be broadly grouped into three categories: (1) those that search for ontologies [12, 15], (2) those that search for individual resources [15, 20], and (3) those that search for concepts that represent a group of individuals [17, 27].Footnote 1 Searching concepts across ontologies represents an ideal granularity middle ground and has applicability in ontology mapping, ontology merging, bootstrapping ontology population, entity annotation, web page classification, and link prediction, all real world applications. With structured content (e.g., knowledge graph) increasing on the web, searching concepts across these is a challenge. In certain domains such as life sciences, there are many overlapping domain ontologies that contain concepts and properties that describe and link concepts. In such a scenario, concept search in itself is a very important task.
To the best of our knowledge, existing concept search approaches can be divided into two types on basis of the nature of the input queries: (1) SPARQL queries [23], which as precise input queries, lead to exact results. However, it requires knowledge of writing SPARQL queries and knowledge of the structure of the ontologies that are to be queried. In reality, learning SPARQL may be an additional burden, and often the structure might not be known to the user. (2) Keyword-based approaches [15, 27], typically use the standard information retrieval techniques such as tf-idf-based and PageRank-inspired algorithms. However, these approaches do not make use of the structure and semantics in ontologies to capture the intents of queries with multiple keywords. In our preliminary work [17] we proposed a concept search framework that only considers relevance. Extending it, in this work, we incorporate diversification of search results and propose a context-based diversification framework that automatically captures fine-grained query intents in the top-k results. We incorporated inferred knowledge using reasoners and refined context further to include annotation properties of widely used vocabularies such as SKOS.
In this paper, we propose a novel keyword-based concept search framework that optimizes both the relevance and diversity of search results. In order to improve search relevance, our framework interprets a query by constructing contexts for concepts from ontology axioms. We exploit the rich and inherent structure and semantics of ontologies and adopt an explicit query interpretation approach [14] in our concept search problem. A keyword query can be ambiguous with multiple intents. Our framework returns the subset of relevant results that contain the most relevant as well as the most diverse results that cover these intents. Our diversification approach achieves the goal of capturing fine-grained intents in the top-k results by using the structure of the ontology.
The technical contributions of our concept search framework are three-fold: (1) the proposal and design of contexts of concepts for their retrieval, (2) explicit, context-based query interpretation based on co-occurrences among keywords in a query, and, (3) explicit, context-based diversification of top-k results using fine-grained search intents.
We have conducted extensive experiments that compare our framework against two concept search systems: the domain-specific BioPortal and the general-purpose Falcons. Our evaluation shows that our framework outperforms both systems by a large margin for both relevance and diversity.
2 Related Work
We relate our work with the broad areas of search approaches and systems.
Semantic Search Approaches. Semantic search engines such as Sindice [32], Swoogle [12, 15], and Watson [11], enable keyword-based search for the ontologies and entities within them. Sindice [32] provides a search interface by using keywords, URI’s and inverse functional properties. Swoogle [12, 15] has developed algorithms to rank the importance of documents, individuals and RDF graphs. The existing semantic search approaches do not leverage the structure and semantics in ontologies to capture the intents of queries with multiple keywords. BioPortal [26] and Falcons [27] are state-of-the-art concept search engines. The Falcons system retrieves concepts, the textual descriptions of which match the keyword query. The system then ranks the results according to the relevance and popularity of the concepts. The BioPortal system provides multiple search functions across ontologies, individuals, and concepts. The BioPortal concept search system is based on the precise or partial matching of the preferred name with the search string. BioPortal use ontology popularity to rank concept search results. We differ in our approach from both these concept search systems in the aspect of searching by using query interpretation and search result diversification techniques.
Indexing and Ranking. SchemEX [24] is an indexing approach for search across the linked open data (LOD) using structured queries. SchemEX consists of three schema layers of RDF classes, RDF types, and equivalence classes with each layer supporting different types of structured queries. However, our framework supports keyword queries and makes use of contexts based on a richer set of ontology constructs. Blanco et al. [5] propose r-vertical index (reduced version of their vertical index) for the RDF entity search problem. The r-vertical index is built by manually categorizing RDF properties in three fields (important, unimportant and neutral). In comparison, our index is built using context information of concepts in the ontologies suitable for our concept search problem. Recent work in the area of Semantic Web resources ranking has largely focused on adapting and modifying the PageRank algorithm. ReConRank [19] is PageRank-inspired [22] algorithm for Semantic Web data. It uses node degree to rank Semantic Web resources in a manner analogous to the PageRank algorithm. ReConRank combines ranks from the RDF graph data sources and their linkage. AKTiveRank [3] ranks ontologies on the basis of how well they cover the specified search terms. The Linked open vocabularies (LOV) [4] search system ranks results on the basis of the popularity of the term in the LOD datasets and in the LOV ecosystem. Butt et al. [6, 7], use offline ranking with the popularity of the concept within the ontology and the popularity of the ontology that contains the concept as the ranking features. Blanco et al. [5] propose instance/entity search using BM25F ranking function. Their ranking function does not exploit proximity information or term dependencies. The existing approaches do not directly exploit the structure in ontologies for indexing and ranking. Dali et al. [9], propose the learning to rank (LTR) [25] approach by using query-independent frequency-based features to rank the results of structured queries. We build the context-based inverted index to interpret the queries, and rank the results of keyword queries using query-based features in the LTR algorithm.
Query Processing and Interpretation. There has been work on structured query processing over LOD and related ontologies. The work on Top-k exploration of query candidates on the (RDF) graph data [31] proposes an intermediate step of converting keyword queries to structured queries. The user needs to select the correct SPARQL query interpretation to retrieve search results. However, our method internally interprets the keyword query without needing to explicitly generate the candidate SPARQL query. Fu and Anyanwu [16] generate query interpretations using the query history as contextual information. However, the queries may not always be iterative and extensive query logs of similar queries may not be available for interpretations. In our approach, we use the context information around a concept across ontologies to interpret the query. We also discuss query interpretation work in the context of implicit and explicit query interpretation. Sawant and Chakrabarti [29] propose implicit generative and discriminative formulations for joint query interpretation and response ranking in keyword-based searches across web documents. Agarwal et al. [1] use probabilistic modeling techniques to mine query templates from query logs for query interpretation. While these approaches may work well for large-scale unstructured data, they may not work in our problem of searching over structured ontologies with low number of redundancies. Our technique of interpreting relations among keywords in the query by using a rich ontology structure is different from the rest of the approaches.
Search Result Diversification. There are two main approaches to diversification: (1) implicit ones that assume that similar documents that cover similar intent/aspects of the query should be demoted to achieve diversified ranking (maximum marginal relevance, or MMR [8]); and (2) those that explicitly model query aspects by sub-queries and maximize the coverage of selected documents with respect to these aspects [10, 21, 28]. These approaches are applied to the unstructured text document search. We believe the diversification techniques have not been designed for the structured data setting of ontologies. Herzig et al. [18] propose language model (LM) approach for consolidating entity search results to reduce redundancy by grouping similar entities. However, their approach does not consider diversity of intent capture in the top-k results. While in our explicit diversification approach, we eliminate redundancy and also capture the fine-grained intents in the top-k search results.
3 Overall Approach
Given a multi-word query Q that consists of m keywords, \(Q = \{k_1, k_2, \ldots ,k_m\}\) on a search space of diverse web ontologies \(\mathcal {O} = \{O_1, O_2, \ldots ,O_n\}\), the goal is to retrieve relevant (named) concepts \(R = \{R_1, R_2, \ldots ,R_p\}\) across these ontologies. We retrieve concept results R by interpreting relations among keywords in the query via the context of a concept (class). Given an ontology, \(O_{g}\) \((g=1,2,\ldots ,n)\), the entities of \(O_g\) include named concepts and named (object-, datatype-, or annotation) properties that are declared in \(O_{g}\). Given an axiom \(a\in O_{g}\) (logical or annotation), let \(\mathtt {sig}(a)\) represent the signature (the set of entities) in a. \(\mathtt {sig}(\cdot )\) is extended naturally to apply to sets of axioms also. For an entity e, let \(\mathtt {annotations}(e)\) represent the values of annotation axioms on e. These annotation axioms include rdfs:label, rdfs:comments, rdfs:isDefinedBy, rdfs:seeAlso as well as those defined in other widely-used vocabularies such as SKOS.
A keyword query is interpreted using the context of each concept. The context of a given concept \(C_{j}\) across ontologies \(\mathcal {O}\) is defined as the set of annotation values of the concept and of the entities that co-occur with \(C_j\) in some axioms in an ontology.
where \(Ax_{C_j}\) and \(Px_{C_j}\) are sets of class-axioms and property-axioms that are relevant to \(C_j\), respectively. Note that \(Ax_{C_j}\) includes subClassOf and EquivalentClasses axioms a that are entailed by an ontology (i.e., \(O \vDash a\)), where both concepts are named concepts (i.e., A and B), and one of them is \(C_j\). These additional, inferred axioms are obtained through reasoning. \(Context(C_j)\) consist of its annotation values (\(\mathtt {annotations}(C_j)\)) and the annotation values of the set of entities that are relevant to \(C_j\) (\(\mathtt {annotations}(e) | e\in \mathtt {sig}(Ax_{C_j}\cup Px_{C_j})\)).
We further employ search result diversification to cover maximum user intents in the top-k search results. We pose our search result diversification problem as a an optimization problem, in which the objective is to maximize the relevance of a result, while minimizing the redundancy among the results. Given a ranked set R of relevant concepts for Q, the goal is to select the subset of concepts \(C_{s}\subseteq R\) that are most relevant to the query and diverse among \(C_{s}\). Along the lines of the MMR [8] framework, our diversification optimization model is:
where \(\mathcal {S}(C_{i})\) is the relevance score of concept \(C_i\), and \(\mathcal {D}(C_{i},C_{s})\) is the diversification score of \(C_i\). \(\mathcal {S}(C_i)\) is obtained by using LTR algorithms. \(\mathcal {D}(C_i, C_s)\) is estimated using the diversity function in which \(C_{i}\) is compared with each of the concepts in \(C_{s}\). The diversity parameter, \(\lambda \in [0,1]\), is the tuning parameter that draws a balance between the relevance and the diversity of a concept.
4 The Concept Search Framework
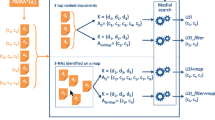
Figure 1 depicts the high-level architecture of our search framework. The components at the bottom are constructed offline, whereas the computations at the top are performed online for each new query. The concept inverted index I is built offline using \(Context(C_j)\), that is relevant to each \(C_{j}\) as defined in Eq. 1 (Sect. 3). Each class and property axiom in \(Ax_{C_j}\) and \(Px_{C_j}\), which is relevant to each \(C_j\) in the ontology corpus is indexed as a field such as rdfs:label, rdfs:comments, rdfs:isDefinedBy. Since the probability of having more than two words together in the ontology corpus is small, we set the shingle size (number of co-occurring words used in co-occurrence computation) to two. In addition, we store the term-vectors for performing co-occurrence computation. We perform natural language processing (NLP) techniques such as tokenization and stemming using the Lucene standard analyzer in order to store the context information in the inverted index.
4.1 The Concept Search Procedure
Given a query Q, the search proceeds by finding concepts with human-readable label \(L(C_j)\) or \(\text {c}{} \textit{lass-name}(C_j)\) (fragment of the URI) that match exactly with the query Q terms as a phrase (line 3 in Procedure CS). We define \(L(C_j)\):
The lexical co-occurrence LC among keywords in a query is evaluated using Pearson’s Chi-squared test (line 6). A Chi-squared value that is greater than 3.841 implies that the keywords co-occur with 95 % confidence. The Pearson’s Chi-squared test returns a set of all co-occurring terms, \(C_{terms}\) in the query. We use \(C_{terms}\) for explicit query interpretation in procedure QI (line 7, further described in Sect. 4.2). QI generates direct and inferred parses for the query using context of concepts. The parses return a set of concepts as search results. Feature vectors (fv’s) are then built for these results to obtain relevance by using LTR model (line 8). An LTR ranking model that is trained offline is applied in order to obtain the relevance score of each search result (line 9, further described in Sect. 4.3). Finally, the search results are diversified to capture fine-grained user intents (line 10, further described in Sect. 4.4).

4.2 Explicit Query Interpretation
Our explicit query interpretation approach generates interpretations by analyzing the interrelationships among the keywords along with the inherently rich structure and semantics of ontologies. Our explicit approach embeds a precise understanding of how each search result is obtained. In the procedure QI, we use the set of co-occurring terms \(C_{terms}\) and all the individual keywords in the query. For each co-occurring terms pair \(C^t \in C_{terms}\), we search for the set of classes CtermClasses for which \(IsExactMatch(L(C_j),C^t)\) is true (line 5). We implement the direct and inferred parse on each of the concepts \(C_j\) in CtermClasses. The direct and inferred parse returns the set of relevant concept results (SearchResults) for the query (line 6–7). If the SearchResults found using direct and inferred parse for co-occurring tokens are less than the threshold (set to 50), we search for a set of classes StermClasses in order to match each keyword \(S^t \in Q\), for which \(IsExactMatch(L(C_j),S^t)\) is true (line 10). We implement the direct and inferred parse on each of the classes \(C_j\) in StermClasses to obtain relevant concept results (SearchResults) for the query (line 11–12). In addition we also return the set of classes CtermClasses and StermClasses as SearchResults if the SearchResults found using direct and inferred parse are less than the threshold (line 14–15).

Direct Parse: The direct parse (DP) returns sets of concepts (SearchResults). DP analyzes the relation among the query keywords by using the context of a concept and is defined as:
where tC is either CtermClasses or StermClasses, and \(sim(Context(C_j), S)\) is calculated using the Jaccard similarity measure.
We explain DP with an example for a query “Myocardial infarction causes” in Fig. 2. The query contains the co-occurring terms pair \(C^t\), “Myocardial infarction”. The keywords “Myocardial infarction” appear as the label of some concept \(C_j\) in our concept index i.e., IsExactMatch \((L(C_j),C^t)=true\). If the context of the same concept \(C_j\) contains “causes” (hence \(sim(Context(C_j), S)>0\) is satisfied), then \(C_j\) with label ‘Myocardial infarction” will be returned as a search result.
Inferred Parse: The inferred parse (IP) returns a set of other concepts that do not directly appear in the query, but rather indirectly through \(\mathtt {SubClassOf}\) or \(\mathtt {EquivalentClasses}\) axioms. The IP is defined as:
where \(OC(C_j)\) is a collection of all other classes that are indirectly related to \(C_j\) through either \(\mathtt {SubClassOf}\) or \(\mathtt {EquivalentClasses}\) axioms, and IP is similarly constructed from classes in \(OC(C_j)\). Here tC, sim are defined in the same way as explained in Eq. 4 of the definition of direct parse.
An example of IP is shown in Fig. 3. Consider the query “Heart attack causes”. The keywords “Heart attack” co-occur and appear as a label of some concept, \(C_j\), in our concept index that is, \(IsExactMatch(L(C_j),C^t)=true\). However, the keyword “causes” is not present in the context of \(C_j\). An equivalent class of \(C_k\), Myocardial_infarction, has the context that contains “causes”. This is interpreted as an inferred relation between “Heart attack” (co-occurring terms) and “causes” (single term) and the class Myocardial_infarction with causes as its relevant property will be returned as a search result.
4.3 Relevance Score Computation
The relevance score for the search results are computed by the ranking model that is built using learning to rank (LTR) algorithms [25]. LTR algorithms are supervised machine learning algorithms. Training data for the ranking model is generated from a query log, in which feature vectors (FV’s) are generated for each combination of query and a result. Components of such FV’s are ranking features, which are obtained using ISUB [30] (12 features) and Jaccard (12 features) similarity, between query terms and concept context fields in the index. The twelve features are computed as the ISUB similarity of query with rdfs:label, rdfs:isDefinedBy, skos:prefLabel, rdfs:comments, rdfs:seeAlso, \(\mathtt {synonym}\), \(\mathtt {dataproperty}\), \(\mathtt {objectpropertydomain}\), \(\mathtt {objectpropertyrange}\), \(\mathtt {SuperClassOf}\), \(\mathtt {SubClassOf}\), \(\mathtt {EquivalentClasses}\), respectively. Similar features are obtained using the Jaccard similarity measure.
The training data of FV’s are used to build LTR models, by employing the RankLib implementationFootnote 2. We use the pairwise RankNet algorithm because the highest normalized distributive cumulative gain (NDCG) value was obtained from this algorithm among the pairwise algorithms by using the query log test data. The RankNet parameters that are used are: the number of epochs to train \(=100\); the number of hidden layers \(=1\); number of hidden nodes per layer \(=10\); and the learning rate \(=0.00005\). The LTR model is trained using 70 % of the log queries in order to learn the weights of the features and 30 % of the queries are used for testing (excluding training queries). The model is then applied to search results in order to obtain the relevance scores for all the concept search results.
4.4 Search Result Diversification
A keyword query may have diverse possible search intents. Search result diversification aims to retrieve k items that are the subset of all relevant results that contain the most relevant and the most diverse intent results. We use the relevance score that is obtained by the LTR algorithm for search result diversification. Search results are diversified by capturing fine-grained query intents explicitly, using the context of concepts.
Baseline Approach. The baseline Implicit approach assumes that similar search results map to the same query intent. Such results should be demoted in order to achieve diversified ranking. Maximum marginal relevance MMR [8] is a canonical technique from the implicit approach. The implicit diversity function is defined as follows:
We calculate the similarity \(SC(C_{i},C_{j})\) among two concepts by comparing their respective context similarity. For example, the context information that is captured in \(\mathtt {sig}(a)\), in which a is a \(\mathtt {SubClassOf}\) axiom of the concept \(C_{i}\) is compared with similar information of the other \(C_{j}\). We use the greedy algorithm [13] by substituting Eq. 6 in Eq. 1 of Sect. 3 in order to implement the implicit diversification.
We iteratively select the best concept result with the highest LTR score (\(S(C_i)\)) from R which can maximize the diversity of the selected concepts \(C_{s}\). The iterative process is repeated until top-k results (\(|C_{s}| = k\)) are obtained.
Fine-Grained Explicit Diversification. Explicit approaches [10, 28] directly map search results to query intents. Diversified ranking is achieved by selecting results that maximize coverage with respect to query intents. Existing explicit diversification approaches obtain query intents from commercial search engines such as Google, which may not be useful in our setting because they are independent of the ontology corpus. Our explicit diversification is based on fine-grained intents that are captured by the contextual information around a concept. We make use of super-class and subclass relations of the returned concepts to generate search intents. Super-classes of concepts cover more generic intents, while subclasses of concepts generate more specific intents.
More specifically, for a query, Q, and the set of most relevant concepts R returned by the LTR model, two levels of intents are generated. Firstly, the top-level intents consists of all of the super-classes of concepts in R. Secondly, the sub-level intents consists of all the subclasses of the super-classes of R.
Top-level intent diversity: The top-level intents are represented as I. Along the lines of the work by Hu et al. [21], we define diversity of the top-level intents as follows:
\(p(C_{i}|x)\), is the probability that \(C_{i}\) satisfies the top-level intent x, and I represents the set of the top-level intents of Q. \(p(C_{i}|x) = sim(L(C_{i}), L(x))\) is estimated as the Jaccard similarity between the labels \(L(C_{i})\) of a concept and the intent L(x), in which \(L(C_{i})\) and L(x) are defined by Eq. 3 in Sect. 4.1.
p(x|Q), which is the probability of x for the given query Q, is estimated by assuming uniform probability distribution \(p(x|Q) = \frac{1}{|I|}\). Uniform intent distribution has been demonstrated to be the most useful [28].
\((1-p(C_{j}|x))\) is the probability that \(C_{j}\) does not satisfy intent x, which indicates that x is less substantially covered and should have higher “priority” in getting more results. The product \(\prod _{C_{j} \in C_{s}} (1-p(C_{j}|x))\) estimates the probability that all concepts \(C_{s}\), that are selected by the LTR model fail to satisfy intent x.
After summing over all query intents, and after being weighted by p(x|Q), the diversity measure in Eq. 8 is the probability that \(C_{i}\) covers the search intents I while the existing list \(C_{s}\), fails to satisfy them.
Sub-level intent diversity: Each of the top-level intents \(x \in I\) is subdivided into sub-level intents S. The sub-level intents are represented as \(S_{x}\) in which x is a top-level intent.
where \(p(C_{i}|y)\) estimates the probabilities that concept \(C_{i}\) satisfies the sub-level intent y, and p(y|Q) is the probability of each of the subclass level intents y for the given query Q. The probability of each of the sub-level intents, y, for query, Q, p(y|Q), is estimated by assuming uniform probability for sub-level intents, \(p(y|Q) = \frac{1}{ \big |I\big | \times \big |S_{x}\big |}\).
By combining the diversity of the top-level and sub-level intents, our fine-grained explicit diversification is estimated as follows
where \(\gamma \) is the tuning parameter for the top-level and the sub-level depending on the granularity of the diversification.
By plugging Eq. 10 into Eq. 1, our diversification optimization model is
where \(\lambda \) is the diversity parameter and \(\gamma \) is the intent parameter for the top-level and sub-level intents.
The model considers the relevance between the concept results \(C_s\) and query Q and the diversity among concepts in \(C_s\). Using a greedy algorithm [13], it iteratively selects the next best concept that is relevant to query Q which maximizes the diversity of selected concepts \(C_{s}\).
5 Evaluation
We compare our system with the search function of two large, widely-used, and openly available ontology repositories, the Bio-medical domain BioPortal,Footnote 3 and the generic FalconsFootnote 4. A summary of the two repositories can be found in Table 1. A separate inverted index was built for each of the BioPortal and Falcons repositories respectively.
We evaluate our system’s performance in terms of relevance only (query interpretation, Sect. 5.1), as well as relevance and diversity (search diversification, Sect. 5.2). Standard information retrieval (IR) ranking measures [25], mean reciprocal rank (MRR) and normalized distributive cumulative gain (NDCG) are used to evaluate our query interpretation approach. The standard search diversification metric of normalized cumulative gain-intent aware (NDCG-IA) [2] is used for evaluation of our diversification technique. Our evaluation dataset is publicly available.Footnote 5
5.1 Query Interpretation Evaluation
Comparison with BioPortal. The BioPortal query log (July 2012 to July 2014) contains more than 2,000 real-world queries as well as click-through data. Among these queries, more than 50 % are multiple-token queries.
Comparison with BioPortal Average Values. We present average NDCG and MRR for our approach vis-a-vis BioPortal for multi-token and single-token queries in Table 2. Our system significantly outperforms BioPortal for multi-token queries, and both systems demonstrate comparable performances in single-token queries.
Query-Wise Comparison with BioPortal. For each query, we calculated the difference between the NDCG values obtained by our system and BioPortal. Of the 2,000 queries, the NDCG values for 1,000 queries (>50 %) are better in our system, and more than 700 queries (>35 %) have the same level of performance. The number of queries in which BioPortal performs better is 300 (<15 %). Our system performs better (>50 %) due to effective use of context information in query interpretation. The level of performance is the same for the queries (>35 %) in which the keywords match the class label exactly. The lower performance (<15 %) may be due to unavailability of context information in the ontologies. The better performance of BioPortal in these (<15 %) queries can be attributed to their statistical consideration of ontology popularity in ranking search results.
Comparison with Implicit Query Interpretation. We evaluated explicit and implicit implementations of query interpretation on the BioPortal dataset. The feature-based implicit model was trained using query logs. Explicit techniques can be more useful in searches over structured data with a low number of redundancies due to the structuredness of the corpora; we also confirm this experimentally. A comparison of the implicit, explicit query interpretation approaches and BioPortal can be found in the Table 3.
Comparison with Falcons. We compared our system vis-a-vis the Falcons search engine [27] in order to explore the generic applicability of our approach. We performed a human-based evaluation in this experiment for better evaluation accuracy and to eliminate noise in automatic clicks. We performed an evaluation on 102 queries that were obtained from two years of TREC web track competitions.Footnote 6 This TREC dataset does not contain single token queries. Our system was evaluated by 30 human users who were undergraduate, graduate, and postgraduate students and had a high level of Web search experiences. Each of the 102 queries was evaluated by at least three of the users. We recorded the binary relevance judgment for the same set of queries for each result on both systems. The performance was evaluated using standard information retrieval measures of NDCG and MRR. Again, our system outperforms Falcons.
Comparison with Average Values of Falcons. We present average NDCG and MRR for our approach vis-a-vis Falcons for multi-token queries in Table 4.
Query-Wise Comparison with Falcons. We calculated the difference of the NDCG and P@k (precision at k) values of our system (with QI) in comparison with Falcons, for the same set of queries. Our NDCG performance was better for >66 % of the queries. It was at par in >25 % and lower in <8 % of the queries. We recorded the top-k (\(k=1,3,5\)) P@k (precision at \(k^{th}\) position) results of our system and Falcons. Of all the queries, the performance of our system in P@1, P@3, and P@5 respectively was better than Falcons in >50 %, >60 %, and >70 % respectively, the same as Falcons in >40 %, >30 %, and >20 % respectively, and lower than Falcons for <10 % for all P@k. The average precision (AP) was calculated by taking an average of P@1, P@3, and P@5 for each query. The positive difference for AP for >70 % queries indicates the better overall performance of our approach.
The subsequent indicative queries give a fair idea of our performance. A query, standard axioms of set theory, has co-occurring keywords set theory and the keywords standard axioms appears in the context of the set theory class. The same query failed to return any results in the Falcons system. For another query, machine learning algorithms, Falcons failed to return relevant results while our system returned relevant result such as machine learning program, and machine learning topic.
5.2 Evaluation of Diversification
Search result diversification was evaluated using a variation of NDCG which is known as the intent-aware normalized cumulative gain measure (NDCG-IA) [2] on the BioPortal and Falcons dataset. We have implemented the implicit diversification approach as defined in Eq. 7 as a baseline, and the explicit diversification approach as defined in the Eq. 11. We set the diversity parameter \(\lambda \) to 0.5, and assigned equal probability to diversity and relevance. Similarly, we set the intent level parameter \(\gamma \) to 0.5, assigning equal priority to top-level and sub-level intents. Table 5 compares the NDCG-IA values produced by our explicit fine-grained diversification method with the implicit diversification baseline, as well as BioPortal and Falcons. Note that separate indices are constructed for the comparison with BioPortal and Falcons.
Comparison with BioPortal. We conducted experiments with 52 queries randomly selected from the BioPortal query log in order to evaluate the effectiveness of intent-capture in our concept search results. Each query was evaluated by three users with a basic level of bio domain expertise and a high level of web search experience. We designed an interface for evaluating our diversification approach. The evaluation interface provided the users with list of intents for search results. The users selected intent for each search result that was used for computing NDCG-IA values.
Comparison of NDCG-IA Values. We report the average NDCG-IA values for the top-10 results of baseline implicit diversification and explicit diversification vis-a-vis BioPortal for multi-token queries in the left part of Table 5. Of the total queries, 70 % fared well with diversification, 25 % were the same as the baseline and 5 % performed lower. The better results for diversification are due to the use to explicit intent capture in our approach. For example, a query Myocardial infarction captures the following intents-myocardial infarction definition, myocardial infarction symptoms, myocardial infarction types, and myocardial infarction causes in the top-k results. On the other hand, the implicit approach removes redundancy but may not address the specific user intents, whereas, the BioPortal captures the intents in their results but not in the top-k.
Comparison with Falcons. We conducted experiments with 50 queries that were randomly selected from the TREC competitions on the Falcons dataset in order to evaluate the effectiveness of intent-capture in our results. Each query was evaluated by three users. The users were graduate students and had a high level of search experience. NDCG-IA was computed for the intents assigned by the user during evaluation.
Comparison of NDCG-IA Values. We present the average NDCG-IA values of baseline implicit diversification and explicit diversification vis-a-vis Falcons, in the right part of Table 5. Of the total queries, 75 % fared well with diversification, 10 % were the same as the baseline and 15 % performed lower. Our explicit diversification techniques effectively captures the fine grained intents. For example, natural language processing applications captures diverse intents such as the linguistic translation process, linguistic topic, and artificial intelligence in our approach, while Falcons returns search results that repeat the single intent in the top-k results.
5.3 Discussion
Our comprehensive log-based and human-based evaluation includes domain-specific and generic ontologies. Our system demonstrated better performance in both settings using standard information retrieval measures, indicating the effectiveness of our framework, especially in multi-token queries. Relation among the keywords in the multi-token queries is effectively captured in our approach. All of our experiments presented in this section (for both multi-token and single-token queries) were found to be statistically significant using the Wilcoxon signed-rank test with p-value <0.0001. Our system’s effectiveness can be attributed to the following factors: (1) Co-occurrence is prevalent among multi-token queries (>50 % queries). (2) Contexts of concepts facilitate effective query interpretation. (3) Our search result diversification methods effectively captures fine-grained intents in top-k results for multi-token queries. As a result, our system seldom returns null or irrelevant results.
6 Conclusion
In this paper we present a novel and effective concept search framework that balances relevance and diversity. We propose to construct contexts for concepts, and use these contexts to (1) interpret user queries and (2) capture fine-grained search intents. The effectiveness of our context-based query interpretation and search result diversification techniques is demonstrated through a comprehensive evaluation against two concept search systems, BioPortal and Falcons. Our evaluation shows that our concept search framework significantly outperforms both systems on widely-used IR metrics.
Our work opens up several directions for further research. Our approach of explicit query interpretation can be improved by incorporating user involvement in the customization of the search. Our explicit diversification formulation can be improved by using proportionality-based optimization techniques. Finally, implementing the applicability of concept search in applications such as ontology population may be useful.
Notes
- 1.
Throughout this paper we use the terms concept and class interchangeably.
- 2.
- 3.
- 4.
- 5.
- 6.
References
Agarwal, G., Kabra, G., Chang, K.C.C.: Towards rich query interpretation: walking back and forth for mining query templates. In: WWW 2010 (2010)
Agrawal, R., Gollapudi, S., Halverson, A., Ieong, S.: Diversifying search results. In: WSDM 2009, pp. 5–14. ACM, New York (2009)
Alani, H., Brewster, C., Shadbolt, N.R.: Ranking ontologies with AKTiveRank. In: Cruz, I., Decker, S., Allemang, D., Preist, C., Schwabe, D., Mika, P., Uschold, M., Aroyo, L.M. (eds.) ISWC 2006. LNCS, vol. 4273, pp. 1–15. Springer, Heidelberg (2006)
Atemezing, G.A., Troncy, R.: Information content based ranking metric for linked open vocabularies. In: SEMANTICS 2014, pp. 53–56 (2014)
Blanco, R., Mika, P., Vigna, S.: Effective and efficient entity search in RDF data. In: Aroyo, L., Welty, C., Alani, H., Taylor, J., Bernstein, A., Kagal, L., Noy, N., Blomqvist, E. (eds.) ISWC 2011, Part I. LNCS, vol. 7031, pp. 83–97. Springer, Heidelberg (2011)
Butt, A.S., Haller, A., Xie, L.: Ontology search: an empirical evaluation. In: Mika, P., et al. (eds.) ISWC 2014, Part II. LNCS, vol. 8797, pp. 130–147. Springer, Heidelberg (2014)
Butt, A.S., Haller, A., Xie, L.: Relationship-based top-K concept retrieval for ontology search. In: Janowicz, K., Schlobach, S., Lambrix, P., Hyvönen, E. (eds.) EKAW 2014. LNCS, vol. 8876, pp. 485–502. Springer, Heidelberg (2014)
Carbonell, J., Goldstein, J.: The use of MMR, diversity-based reranking for reordering documents and producing summaries. In: SIGIR 1998 (1998)
Dali, L., Fortuna, B., Duc, T.T., Mladenić, D.: Query-independent learning to rank for RDF entity search. In: Simperl, E., Cimiano, P., Polleres, A., Corcho, O., Presutti, V. (eds.) ESWC 2012. LNCS, vol. 7295, pp. 484–498. Springer, Heidelberg (2012)
Dang, V., Croft, W.B.: Diversity by proportionality: an election-based approach to search result diversification. In: SIGIR 2012, pp. 65–74. ACM, New York (2012)
d’Aquin, M., Motta, E.: Watson, more than a semantic web search engine. Semant. Web 2(1), 55–63 (2011)
Ding, L., Pan, R., Finin, T.W., Joshi, A., Peng, Y., Kolari, P.: Finding and ranking knowledge on the semantic web. In: Gil, Y., Motta, E., Benjamins, V.R., Musen, M.A. (eds.) ISWC 2005. LNCS, vol. 3729, pp. 156–170. Springer, Heidelberg (2005)
Drosou, M., Pitoura, E.: Search result diversification. SIGMOD Rec. 39, 41–47 (2010)
Fagin, R., Kimelfeld, B., Li, Y., Raghavan, S., Vaithyanathan, S.: Understanding queries in a search database system. In: PODS 2010, pp. 273–284 (2010)
Finin, T., Peng, Y., Scott, R., Joel, C., Joshi, S.A., Reddivari, P., Pan, R., Doshi, V., Ding, L.: Swoogle: a search and metadata engine for the semantic web. In: CIKM 2014 (2004)
Fu, H., Anyanwu, K.: Effectively interpreting keyword queries on RDF databases with a rear view. In: Aroyo, L., Welty, C., Alani, H., Taylor, J., Bernstein, A., Kagal, L., Noy, N., Blomqvist, E. (eds.) ISWC 2011, Part I. LNCS, vol. 7031, pp. 193–208. Springer, Heidelberg (2011)
Gavankar, C., Li, Y.F., Ramakrishnan, G.: Context-driven concept search across web ontologies using keyword queries. In: K-CAP 2015, pp. 20:1–20:4. ACM (2015)
Herzig, D.M., Mika, P., Blanco, R., Tran, T.: Federated entity search using on-the-fly consolidation. In: Alani, H., et al. (eds.) ISWC 2013, Part I. LNCS, vol. 8218, pp. 167–183. Springer, Heidelberg (2013)
Hogan, A., Harth, A., Decker, S.: Reconrank: a scalable ranking method for semantic web data with context. In: 2nd Workshop on Scalable Semantic Web Knowledge Base Systems (2006)
Hogan, A., Harth, A., Umbrich, J., Kinsella, S., Polleres, A., Decker, S.: Searching and browsing linked data with SWSE: the semantic web search engine. J. Web Semant. 9(4), 365–401 (2011)
Hu, S., Dou, Z., Wang, X., Sakai, T., Wen, J.R.: Search result diversification based on hierarchical intents. In: CIKM 2015, pp. 63–72. ACM (2015)
Kleinberg, J.M.: Authoritative sources in a hyperlinked environment. J. ACM 46, 604–632 (1999)
Kollia, I., Glimm, B., Horrocks, I.: SPARQL query answering over OWL ontologies. In: Antoniou, G., Grobelnik, M., Simperl, E., Parsia, B., Plexousakis, D., De Leenheer, P., Pan, J. (eds.) ESWC 2011, Part I. LNCS, vol. 6643, pp. 382–396. Springer, Heidelberg (2011)
Konrath, M., Gottron, T., Staab, S., Scherp, A.: Schemex - efficient construction of a data catalogue by stream-based indexing of linked data. J. Web Sem. 16, 52–58 (2012)
Liu, T.Y.: Learning to Rank for Information Retrieval. Springer, Berlin (2011)
Noy, N.F., Alexander, P.R., Harpaz, R., Whetzel, P.L., Fergerson, R.W., Musen, M.A.: Getting lucky in ontology search: a data-driven evaluation framework for ontology ranking. In: Alani, H., et al. (eds.) ISWC 2013, Part I. LNCS, vol. 8218, pp. 444–459. Springer, Heidelberg (2013)
Qu, Y., Cheng, G.: Falcons concept search: a practical search engine for web ontologies. IEEE Trans. Syst. Man Cybern. Part A 41(4), 810–816 (2011)
Santos, R.L., Macdonald, C., Ounis, I.: Exploiting query reformulations for web search result diversification. In: WWW 2010, pp. 881–890. ACM, New York (2010)
Sawant, U., Chakrabarti, S.: Learning joint query interpretation and response ranking. In: Proceedings of the 22nd International Conference on World Wide Web (2013)
Stoilos, G., Stamou, G., Kollias, S.D.: A string metric for ontology alignment. In: Gil, Y., Motta, E., Benjamins, V.R., Musen, M.A. (eds.) ISWC 2005. LNCS, vol. 3729, pp. 624–637. Springer, Heidelberg (2005)
Tran, T., Wang, H., Rudolph, S., Cimiano, P.: Top-k exploration of query candidates for efficient keyword search on graph-shaped (RDF) data. In: ICDE 2009, pp. 405–416 (2009)
Tummarello, G., Delbru, R., Oren, E.: Sindice.com: weaving the open linked data. In: Aberer, K., et al. (eds.) ASWC 2007 and ISWC 2007. LNCS, vol. 4825, pp. 552–565. Springer, Heidelberg (2007)
Acknowledgments
We thank Prof. Mark Musen and Prof. Paul Alexander of Stanford University for sharing the BioPortal query log. We would like to thank Prof. Gong Cheng for his advice and guidance on working with Falcons.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Gavankar, C., Li, YF., Ramakrishnan, G. (2016). Explicit Query Interpretation and Diversification for Context-Driven Concept Search Across Ontologies. In: Groth, P., et al. The Semantic Web – ISWC 2016. ISWC 2016. Lecture Notes in Computer Science(), vol 9981. Springer, Cham. https://doi.org/10.1007/978-3-319-46523-4_17
Download citation
DOI: https://doi.org/10.1007/978-3-319-46523-4_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46522-7
Online ISBN: 978-3-319-46523-4
eBook Packages: Computer ScienceComputer Science (R0)