Abstract
Traditionally, in health surveillance, high risk zones are identified based only on the residence address or the working place of diseased individuals. This provides little information about the places where people are infected, the truly important information for disease control. The recent availability of spatial data generated by geotagged social media posts offers a unique opportunity: by identifying and following diseased individuals, we obtain a collection of sequential geo-located events, each sequence being issued by a social media user. The sequence of map positions implicitly provides an estimation of the users’ social trajectories as they drift on the map. The existing data mining techniques for spatial cluster detection fail to address this new setting as they require a single location to each individual under analysis. In this paper we present two stochastic models with their associated algorithms to mine this new type of data. The Visit Model finds the most likely zones that a diseased person visits, while the Infection Model finds the most likely zones where a person gets infected while visiting. We demonstrate the applicability and effectiveness of our proposed models by applying them to more than 100 million geotagged tweets from Brazil in 2015. In particular, we target the identification of infection hot spots associated with dengue, a mosquito-transmitted disease that affects millions of people in Brazil annually, and billions worldwide. We applied our algorithms to data from 11 large cities in Brazil and found infection hot spots, showing the usefulness of our methods for disease surveillance.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
There is an increasing availability of geolocated data generated by mobile phones, connected vehicles and geotagged social media, among other sources. This is enabling a broad spectrum of applications and services that exploit such data and demand the development of novel data mining models and algorithms that support those tasks. Building such models and algorithms require that we are able to handle novel types of data, such as user’s movement record as well as the noisy and the incomplete nature of the data. We may glance the problem complexity by checking the right plot of Fig. 1, which depicts the movement of Twitter users in 2015 in a Brazilian city, Rio de Janeiro (each line segment shows a user’s movement – location change – between two consecutive messages).
In this paper we tackle one of these disruptive application scenarios: determining infection hot spots, that is, the high risk zones where people got infected by a disease. Our proposal adopts the case-control framework, where, by contrasting the case and control individuals’ characteristics, we learn about the disease dissemination process. The input is composed of trajectories, which are sequences of user locations that provide an estimate of the users’ movements as they drift on the map. We depict this application scenario in the left plot of Fig. 1, where each polygonal line is a trajectory that represents either a case (red) or a control (blue) individual, and we want to determine whether there are regions (represented by the shaded area in the left plot of Fig. 1) where infection is more likely, manifested by a larger number of case trajectories than control trajectories, among other evidences. Such information may be key to surveillance and disease mitigation actions.
Although the main idea seems simple, there are a large number of challenging data mining issues that require the development of novel models and algorithms to the problem. At first, this task resembles spatial cluster detection, which aims at detecting localized spatial regions or zones, called spatial clusters, where the likelihood of some event occurrence is higher than in the rest of the map [8, 11, 17]. There have been several different proposals for detecting spatial clusters, but all of them are based on the same premise: each entity is associated with one or at most two locations. Thus, our proposal differs significantly from current spatial cluster detection strategies in the sense that there is no limit on the number of events, and then locations, associated with a person. Further, there are two other characteristics that make our problem more challenging: (i) the number of events per trajectory may vary significantly, and (ii) we do not know in advance which events represent the actual infection.
In this paper we propose two stochastic models with their associated algorithms to mine this new type of data. The Visit Model finds the most likely zones that a diseased person visits, while the Infection Model finds the most likely zones where a person gets infected while visiting. To the best of our knowledge this is the first work that goes beyond predicting disease incidence rate from social media data. Our approach leverages the geo-tagged social media messages in order to discover potentially high infection risk zones. Specifically our contributions are as follows:
-
We describe the problem of detecting infection hot spots from trajectory data in a case-control framework (Sect. 2).
-
We propose two novel models, and the respective algorithms, the Visit Model and the Infection Model, for the discovery of significant infection hot spots. Our algorithms address all three aforementioned issues (Sect. 3).
-
We propose an extraction and modeling strategy of Twitter data to the hot spot detection problem in the context of dengue (Sect. 4).
-
We present detailed experimental results to illustrate our approach in action by applying our algorithms to a set of 11 Brazilian municipalities analyzing more than 100 million tweets issued in 2015 (Sect. 5).
2 Problem Description
Social media data represent a rich and promising source of plenty, cheap, and timely data that has been only tapped in its usefulness. The excitement involving the use of social media as a social sensor could be felt by the countless number of research works using this kind of data [6, 16, 18]. In our case, we are interested in probing the usefulness of social data spontaneously generated by users as a way to identify the location, shape, and size of high risk zones and to determine its statistical significance. Depending on the application, we believe that these data may be more precise in the detection of such hot spots than other more standard data and, in many cases, they may be the only data available. Indeed, this latter observation is exactly the case of dengue surveillance (see Sect. 4.1), since there is scarce, if any, information about the place where people are being infected with dengue by the transmitter mosquito. As dengue usually is a debilitating disease that causes much pain, our assumption is that infected individuals will report what they are experiencing in social media [6, 19].
In this work we use dengue and Twitter to instantiate our proposal, but it is obviously general and can be applied to a large range of other situations (see Sect. 7). Each user in the database is classified either as a case or a control individual. The separation of cases and controls is based on the content of tweets text: users mentioning personal experience with dengue are labelled as cases, otherwise, they are labelled as controls. In the left plot of Fig. 1, we have \({N=6}\) cases and \(M=4\) controls identified by the red and blue polygonal lines, respectively. The vertices of the polygonal lines correspond to the locations of tweets issued by each individual. The tweets of a single individual are connected in chronological order and hence we refer to the polygonal lines as trajectories. For the cases, the specific dengue-labelled messages are marked by a hatched ground area. We also show in the same figure a candidate hot spot Z (shaded area), a spatial zone potentially riskier than other regions in the map.
The mining task is to scan the map varying the position, shape, and size of the candidate zones, looking for the zone \(\hat{Z}\) that most likely is a higher risk area. After finding this most likely hot spot \(\hat{Z}\), we calculate its probability of occurrence to evaluate whether there is enough evidence to call it a real cluster. The simple schematic illustration is put in due perspective when we look at the right-hand map in Fig. 1. The large amount of data and the impossibility to visually identify any meaningful pattern supports the demand for new data mining models and algorithms.
Usual approaches [8, 11, 17] for spatial cluster detection can not be used here. All spatial detection methods have a single location associated with each case or control individual, usually their residential addresses or working places. In our case, we have a completely different spatial data structure. First, each i-th individual is not associated with a single location, but with a series of \(n_i\) successive positions \(\mathbf {x}_i\) in the map. There is no single unambiguous position to assign each case or control but rather a sequence of positions. Usual methods are not able to handle this scenario.
Second, the number \(n_i\) of positions of each individual is quite variable. For some individuals, \(n_i\) is small, with less than 10 positions. Others may contribute a large number of positions, reaching more than 100 tweets. Clearly, the locations can not be put on the map ignoring the variable contribution of each individual. To make this point clearer, imagine an extreme situation where 3 case individuals contribute each one with two positions, one in a risky zone, and another one outside. At the same time, an additional case individual has 200 tweets spread all over the map. This extreme individual would dominate the analysis if we do not take the sample size \(n_i\) into account. Again, this is not considered in the usual techniques, where each individual contributes with a single point.
Third, and more challenging, the positions of the dengue-labelled tweets are not necessarily those where the infection risk is higher. Indeed, our assumption is that the individual entire trajectory (and not a single position) will be informative of the risk areas. Someone affected by dengue could tweet about his condition days after recovery and at a location not associated with its infection place. This challenge is addressed through sampling the controls. We expect that contrasting between the spatial pattern of trajectories of the case and the control individuals, riskier zones should be pinpointed by our algorithms.
3 Mining for Hot Spots from Trajectories
As mentioned, we adopted a case-control framework, where the data consist of locations, within a specified geographical region, of all known cases of a particular disease, and of a random sample of controls drawn from the population at risk. Each individual carries a set of features corresponding to known or hypothesized risk-factors for the disease in question.
In our analysis, the key innovation is that the input is a series of locations rather than a single location for each individual. As in a standard case-control study, each sampled person is classified either as a dengue case or a non-dengue (control) individual. We labelled the individuals such that the first N of them are the cases and the last M are the controls. Let \(\mathbf {x}_i = (x_{i,1}, \ldots , x_{i,n_i})\) be the point events associated with the \(n_i\) tweets issued by the i-th individual, \(i=1, \ldots , N+M\). Each \(x_{i,k}\) represents the geographical tweet location such as a latitude-longitude coordinate pair. For the cases \(i=1, \ldots , N\), at least one tweet in \(\mathbf {x}_i\) refers to a personal dengue experience and their specific locations will be denoted dengue-labelled tweets hereafter. Typically, there will be a small percentage of dengue-labelled tweets for each individual. None of the control individual tweets are dengue-labelled.
Let \(\mathcal {Z}\) be a (large) set of geographical zones that are candidates to be infection hot spots. The left plot of Fig. 1 helps us to describe how our algorithm works. There are potentially infinite zones in \(\mathcal {Z}\) and they cover the entire region under analysis. By varying \(Z \in \mathcal {Z}\) we scan the map looking for the zone \(\hat{Z}\) that most likely is a higher risk area. After finding this most likely hot spot \(\hat{Z}\), we calculate its likelihood to evaluate whether there is enough evidence to identify it as a hot spot. Secondary clusters are also searched, as we explain later.
Our approach is to contrast the number of cases and controls visiting the potential zone. With a meaningful contrasting score, we should then scan the map to find the most likely zone. We considered two different probability scores, depending on how we calculate conditional probabilities of relevant events. In the first, we use the probability that someone visits the candidate zone Z given that she is either a case or a control individual. Intuitively, a risky zone Z should have this visit probability higher for cases than for controls. This first approach is called the Visit Model. In the second, we use the probability that someone gets infected given that it visits the candidate zone Z. Intuitively, we anticipate that cases visit Z more often than controls. This second approach is called the Infection Model. We present them formally next.
3.1 Visit Model
Let \(V_{i,z}\) be the random number of tweets in Z among the \(n_i\) total number of tweets issued by the i-th individual. Use \(\mathbbm {1}{[A]}\) to represent the indicator random variable that the event A occurs. Hence \(\mathbbm {1}{ [ V_{i,z} \ge 1 ] }\) is the binary random variable indicating whether the i-th individual ever tweeted inside the candidate zone Z. These random variables can be assumed independent, but they are not identically distributed as the success probability depends on the number \(n_i\) of tweets issued by each individual. Denote by \(p=p(Z)\) the probability that, giving that a case individual is tweeting, she does it from within Z. Let \(\bar{p}=\bar{p}(Z)\) be the similar probability for a control individual. We are interested in zones where \(p(Z) > \bar{p}(Z)\).
For a user who is a case, we have \(\mathbb {P} \left( V_{i,z} \ge 1 \right) \) equals to \(1 - (1-p)^{n_i} \) and, for a control user, it is equal to \(1 - (1-\bar{p})^{n_i} \). Considering a fixed zone Z, the visit model likelihood for the observed \(N+M\) binary indicators \(\mathbbm {1}{ [ V_{i,z} \ge 1 ] }\) is given by
Let \(N(\bar{Z}) = \sum _{i=1}^N n_i \mathbbm {1}{[V_{i,z} = 0]}\) and \(M(\bar{Z}) = \sum _{i=N+1}^{N+M} n_i \mathbbm {1}{[V_{i,z} = 0 ]}\) be the total number of tweets from users (both cases and controls) who did not visit zone Z, respectively. Hence, the log-likelihood \(\ell _1(Z, p, \bar{p}) = \log (L_1(Z, p, \bar{p}) )\) for this first model can be written as
3.2 Infection Model
We will estimate the probability that someone issues a dengue-labelled tweet (and becomes a case) given that she visited k times the region Z. Let \(r = r(Z)\) be the infection risk inside the candidate cluster and \(\bar{r} = r(\bar{Z})\) the infection risk in \(\bar{Z}\), the region outside Z. We are interested in zones Z where \(r(Z) > r(\bar{Z})\).
Let \(I_i\) be the binary indicator that the individual i is a case. We assume that these binary random variables are independent. They are not identically distributed since their probability of \(I_i=1\) depends on the number of visits \(V_{i,z}\) by the i-th individual to the zone Z. We have
Therefore, the likelihood of the pattern of cases and controls is given by
and therefore the log-likelihood expression is given by
3.3 Evaluating the Data Evidence
Recall that \(\mathcal {Z}\) is the set of candidate zones to be scanned. The test statistic we adopt for the Visit Model is
and an analogous formula defines \(T_2\) for the Infection Model. In order to verify its statistical significance, we must use Monte Carlo simulation to obtain the null hypothesis distribution of \(T_1\) and \(T_2\) as the exact or asymptotic analytic calculation is not feasible. The null hypothesis is given by either \(H_0: p = \bar{p}\) or \(H_0: r = \bar{r}\) for all \(Z \in \mathcal {Z}\) for the Visit Model and the Infection Model, respectively.
The Monte Carlo distribution is determined by randomly permuting the labels of cases and controls among all individuals. Using this pseudo dataset, we proceed the entire scan over all \(Z \in \mathcal {Z}\) to obtain a pseudo value for \(T_1\) and \(T_2\). As this will be replicated several times, we call these values \(T_1^{(1)}\) and \(T_2^{(1)}\). We then select another random permutation of the labels, scan the zones and find \(T_1^{(2)}\) and \(T_2^{(2)}\). Independently, we repeat this procedure a large number \(B-1\) of times generating a set of pseudo values plus the values calculated with the actually observed dataset: \(T_1, T_1^{(1)}, T_1^{(2)}, \ldots , T_1^{(B-1)}\) and \(T_2, T_2^{(1)}, T_2^{(2)}, \ldots , T_2^{(B-1)}\). Under the null hypothesis, these values are independent and identically distributed. Therefore, the rank of the real observed statistics \(T_1\) and \(T_2\) are uniformly distributed on the integers \(1, \ldots , B\). This implies that an exact p-value for the null hypothesis of each model is given by
and
The test is significant at the level \(\alpha \in (0, 1)\) if \(p_m < \alpha \). When either test is significant, the most likely zone is given by the corresponding maximizing argument \(\hat{Z}\) in (4).
We also identify secondary clusters, zones with highly significant p-values, which do not intersect with the most likely zone \(\hat{Z}\). The non-intersecting restriction is necessary because, if one zone \(\hat{Z}\) is the most anomalous in \(\mathcal {Z}\), many other sets in \(\mathcal {Z}\) that are only slightly different from \(\hat{Z}\) will produce very similar likelihood numbers. These zones should be ignored since the most anomalous among them has already been pinpointed. Among the non-intersection zones, we look for those whose p-value \(p_m\) is smaller than \(\alpha \) where the p-values are calculated as described above.
3.4 Contrasting the Two Models
In this section, we discuss in more detail the two proposed models aiming at providing an understanding of the differences between them. In particular, we want to distinguish between the two approaches in an intuitive way and hence explain when and how we can have one of the models detecting a certain hot spot while the other model is insensitive to this same cluster presence.
Avoiding the rigorous mathematical notation, let us define two random events. The first one is denoted by C and represents the random selection of an individual from the database that is dengue-affected or simply a case. Its complementary event is \(\bar{C}\) and represents the selection of a control individual. Given that a tweet is posted by a user, we denote by \(W_Z\) the event that it is issued from Z while \(W_{\bar{Z}}\) means that it is from outside Z.
The visit model considers two conditional probabilities, \(p = \mathbb {P}( W_Z | C)\) and \(\bar{p} = \mathbb {P}(W_Z | \bar{C})\), while the infection model considers the corresponding inverse conditional probabilities, \(r = \mathbb {P}(C | W_Z)\) and \(\bar{r} = \mathbb {P}(C | W_{\bar{Z}} )\). Intuitively, the visit model scans the map looking for a zone Z where p and \(\bar{p}\) are quite different. The infection model searches for a zone where the difference between r and \(\bar{r}\) is large. They can find distinct and separate zones in this process. The main reason is the usual large difference we find between conditional probabilities \(\mathbb {P}(A|B)\) and \(\mathbb {P}(B|A)\) of events A and B. The connection between the two is given by the Bayes rule: \(\mathbb {P}(A|B) = \mathbb {P}(B|A) \mathbb {P}(A)/\mathbb {P}(B)\). Since the factor \(\mathbb {P}(A)/\mathbb {P}(B)\) is the link between the two, when we have very different values for \(\mathbb {P}(A)\) and \(\mathbb {P}(B)\) we can expect large differences on the two directions for the two conditional probabilities, \(\mathbb {P}(A|B)\) and \(\mathbb {P}(B|A)\).
This is indeed what one can expect in our dengue application. The unconditional probabilities \(\mathbb {P}(C)\) and \(\mathbb {P}(W_Z)\) are typically very different. As we take about 3 times more controls than cases, we anticipate \(\mathbb {P}(C) \approx 1/4\). For a localized zone Z, even if it is highly infectious, we should not expect \(\mathbb {P}(W_Z) > 1/20\). Hence, zones detected by one of the models should not be predicted as the likely output by the other model.
An additional enlightening way to contrast the two models is to consider the extreme situation in which each user has issued a single tweet (that is, \(n_i =1\)). As a consequence, \(k_i\) is equal to 0 or 1 and the likelihood for the two models may be considerably simplified. Remember that \(N(\bar{Z})\) is the number of tweets from cases posted from outside Z (or \(\bar{Z}\)) while \(M(\bar{Z})\) is the analogous count for the controls. The notation N(Z) and M(Z) has the obvious definition: counts of tweets from inside Z.
For the visit and infection models, their respective log-likelihood functions \(\ell ^{'} _1 = \ell _1(Z, p, \bar{p})\) and \(\ell ^{'} _2 = \ell _2(Z, r, \bar{r}) \) are reduced to
These likelihood functions for this extreme situation show that the two models use the data differently to search for suspicious zones Z. They both point out to likely high infection risk areas but they use different approaches in the process and may spot different potential candidates. The two approaches are logically consistent and produce meaningful results. They are complementary to each other and should not be seen as opposites.
3.5 Spatial Scan Statistics as a Particular Case
Expression (6) shows that the usual spatial scan statistic [8, 11, 17] is a particular case of our infection model. Assuming that each sampled individual has a single spatial location (usually her residential address), the notation r represents now the probability that she is a disease case given that she is within Z. The probability \(\bar{r}\) is the same probability for someone living outside Z. Then, (6) is the Bernoulli likelihood used by the original spatial scan statistic. That is, when there is a single location for each individual, we obtain the classic spatial scan statistic by applying our infection model.
4 Case Study: Dengue in Brazil
In this section we present the motivation behind our evaluation scenario, dengue disease surveillance in Brazil. Also, we describe the Twitter data collection process and how we properly filter the data in order to obtain the case-control individuals’ trajectories.
4.1 Context
Despite all the progress achieved in the twenty-first century, diseases transmitted by insects are still challenging our health services and policy makers. The recent outbreak of Zika virus in Brazil and other Latin American countries, potentially associated with thousands of microcephalic birth cases, prompted The World Health Organization (WHO) to declare the Zika Infection a world health threatFootnote 1. Other disease that is transmitted by the same mosquito, Aedes aegypti, is dengue.
With an estimated 50–100 million infections globally per year [3], dengue is currently regarded as the most important mosquito-borne viral disease. Dengue affects over 100 endemic countries in tropical and sub-tropical regions of the world, mostly in Asia, the Pacific Region and the Americas. Presenting four distinct viral serotypes, dengue fever may range from severe flu-like illness up to a potentially lethal complication known as severe (or hemorrhagic) dengue. The World Health Organization estimates that 3.9 billion people are at risk of infection with dengue viruses. However, the true impact of the disease is, sometimes, difficult to assess due to misdiagnosis and underreporting [2]. Global dengue incidence still grows in number and severity of cases and also in the amount of new affected areas. This is most due to modern climate changes and to socioeconomic, and viral evolution [12]. However, the potential drivers of dengue are often difficult to detect and factor out. Since there is no current approved vaccine to protect the population against the virus [12], epidemiological surveillance and effective vector control are still the mainstay of dengue prevention.
Dengue is a serious concern in Brazil. In 2015, more than US$ 300 million were spent in surveillance and prevention actionsFootnote 2. This is a significant figure for Brazilian standards and, despite its magnitude, more than 1.6 million cases were recorded in 2015. This number represents a rate of 813 cases per 100 thousand inhabitants, well above the redline indicated by the WHO (300 cases).
Most studies for diseases such as dengue place the cases at individuals’ residential addresses, which may quite often not be the infection location. The relatively easy to obtain residential address may be a poor indicator of the zones where humans and infected mosquitoes tend to meet each other. These zones are hard to determine, since the necessary information about them is scarcely available. Indeed, such information comprises data on the mosquito prevalence, its infection rate, and the human movement in each potential zone. Notwithstanding the task difficulty, identifying the most risky places would be invaluable because we could focus the expensive and diffuse preventive efforts undertaken until now.
4.2 Data Acquisition and Preprocessing
The data used in our experimental analysis were acquired through the Twitter streaming application programming interface (API) [1], using a geographic boundary box that covers the whole Brazilian territory. Consequently, all collected tweets are geo-tagged with lat/long GPS coordinates. The collecting period comprises from January 1st, 2015 to December 31th, 2015. During this time we were able to collect 106,784,441 tweets comprising a multitude of subjects. We want to use this data to search for zones that increase the likelihood that an initially control individual becomes a case.
Since the majority of users usually moves within the same city, we decided to perform our analysis at the city level. This granularity is also interesting because, in Brazil, the decision process regarding dengue surveillance actions is under the responsibility of each city hall. Thus, a fine geographic scale analysis would lead to focused preventive efforts. Since the messages are geocoded, to obtain the data from a specific city is straightforward. The Twitter API provides the location based on the lat/long coordinates. We use the assigned location by filtering the corresponding tweet field. We choose 11 municipalities (see Sect. 5 for the explanation) to analyze. Table 1 summarizes the data for each selected city.
For each city analyzed, we filtered the data indicating whether the user is a case individual. We defined the keywords dengue and aedes, and started a search throughout the data. Previous works showed a high correlation between official dengue reports and Twitter data collected with such keywords [6, 19]. We also check for misspelling and ignore letter case. Since the vocabulary in text-based social media is very dynamic, the retrieved messages based on keywords may not be actually associated with people reporting personal experience with the disease. Hence, we classified the messages according to the sentiment expressed in the textual content. To classify the messages, we preprocessed texts by filtering out accents marks and URL’s. Bi-grams were created by joining adjacent words with a separator, and stop-words were removed as well as bi-grams composed of two stop-words. The classification was performed in a supervised manner. We manually labelled a set of tweets from a different Twitter collection specifically about dengue disease. This collection is performed based on the same keywords. Similar to [6, 19], the tweets were classified into one out of five categories: Personal Experience, Information, Opinion, Campaign and Irony/Sarcasm, using the the Lazy Associative Classification algorithm (LAC) [20]. Next, we separated the messages assigned to the Personal Experience category, since they may indicate a closer relationship between the user and the disease. These messages represent the dengue-labelled tweets for the case individuals.
4.3 Case-Control Trajectories
Recall that, each user in the database is classified as either a case or a control individual, and the separation of cases and controls is based on the content of tweets text, as described above. Then, for each city we build the case-control trajectories as follows.
Case-trajectories. In order to build the case individuals trajectories we started by separating all unique users who posted a dengue-labelled message. Then, we retrieved all other tweets sent by these users. For each case individual, her list of messages composes the trajectory. Such strategy is interesting because we are implicitly considering that the users must have been infected at some point in their daily movements and not exactly where the dengue-labelled messages were sent. After that, we excluded highly active users to avoid, for instance, bots. We adopted a 5-message-per-day threshold, which represents a maximum of 1825 messages per year. The users with total number of messages above this threshold are excluded from the dataset.
Control-trajectories. The control individuals group comprises all users who never posted a message containing any of the keywords used to define the case individuals group. Therefore, none of the control individuals tweets are dengue-labelled. We defined the same threshold to exclude highly active users. The number of control individuals is much larger than the number of case individuals. Thus, we sampled the control individuals. We stratified the case individuals according to the total number of messages in ranges of 10. Then, for each range we sampled the number of control users as 3 times the number of case users in that same range. When the number of control users in a given range was not enough to reach the amount required, we used the total available.
5 Experimental Analysis
After generating the dataset for each selected city as described in the previous section, we proceeded to the experimental analysis. For each one of the 11 selected cities (see Table 1) we applied the Visit Model and the Infection Model to search for infection hot spots. Among the selected cities we included 7 state capitals (Belém, Belo Horizonte, Curitiba, Goiânia, Natal, Rio de Janeiro and São Paulo) with at least one capital from a major Brazilian region. We also decided to assess our models using data from municipalities facing high epidemics bursts. Therefore, we included 4 other cities: Campinas, Limeira, São José dos Campos and Sorocaba. For instance, while in 2014 Sorocaba reported less than 400 dengue cases, in 2015 the same city reported more than 50 thousand cases.
In order to run the algorithms, the zones Z are defined by overlaying different grids on the map and each grid cell corresponds to a zone to be scanned. The size of the grid cells vary in order to accommodate risk zones that present different characteristics. We set the number of Monte Carlo replicas to \(B-1 = 999\) and define the significance level as \(\alpha = 0.05\). Among the 11 selected cities, in 4 of them at least one of the models was able to find one or more significant hot spots. Table 2 summarizes the results.
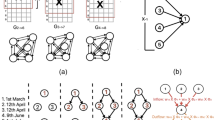
First of all, we point out that our models were able to find infection hot spots in 3 cities that faced the aforementioned strong surges. Despite the significance level being \(\alpha = 0.05\), we considered the borderline region found in SJ. Campos as significant. In the context of disease surveillance, it would be also important to check such zones. We observe that, in Goiânia and Limeira, the zones pinpointed by the Visit Model were visited by most of the case individuals, since the Visit Model searches for the most likely zones where case individuals visit. On the other hand, the zones identified by the Infection Model comprise a lower number of case individuals seeking for more restricted areas. In fact, the size of the zones found by the models differ. The Visit Model usually finds larger regions whilst the Infection Model finds smaller regions. Figure 2 depicts the zones found by each model in the corresponding cities. Notice that in Limeira the models identified different regions within the same city. These results also point out the complementarity of the models, so that they may be used together towards establishing two different levels of surveillance.
Maps of the cities with the hot spots found by both models. The cities are Goiânia, Limeira, São José dos Campos and Sorocaba. The green and black squares depict the zones found by the Visit and Infection models respectively. We also display the case and control individuals trajectories as red and blue points, respectively. (Color figure online)
After we find the significant zones, we may analyze them in detail to observe their characteristics. We show this more detailed analysis for Goiânia. Figure 3 displays a zoom in the zone identified by the Visit Model and the respective case-control trajectories. We point out that there are many places, such as, college campi, hospitals and parks inside the zone. Since those places are non-residential, current techniques would never consider them as potential infection hotspots, in the face of a rise in the number of cases. This is another interesting feature of our algorithms, they can point out places which represent a better approximation of where people might have been infected, being worthy to investigate those areas.
6 Related Work
Spatial cluster detection is a special class of data mining problem within the more general anomalous pattern detection problem. The assumed structure of the input data is a spatial point location, such as latitude-longitude pair, besides the usual features associated with each of them. The seminal paper [7] originated a flow of work and its large impact may be explained by a breakthrough contribution. They developed a practical way, the spatial scan statistics, to take into account the multiple testing involved in the search of anomalous regions. They showed how a simple Monte Carlo reference distribution could be obtained from the data and how it controls the false positive level of the potentially infinite statistical tests involved. This idea opened the door to many additional developments [4, 5, 9, 13–15, 17, 21, 24]. While the recent availability of spatial data offers a unique opportunity, the existing data mining techniques for spatial cluster detection fail to address this new setting as they require a single location to each individual under analysis.
On the other hand, there has been fruitful research exploiting spatial data for a variety of purposes, such as, discovering the spatial dependency of objects [22], understanding mobility patterns [10] and clustering similar trajectories [23], to name a few. However, none of the strategies proposed so far focused on searching for hot spots by contrasting trajectory data of targeted populations with those from control populations as we have done here. In this sense, this paper has a two-fold contribution. First, it generalizes the spatial cluster detection approaches by considering the individual trajectory data instead of a single point. Second, it describes the aforementioned problem in the context of disease surveillance and proposes two algorithms to mine the data.
7 Concluding Remarks
Exploiting the large amount of available data for addressing relevant social problems has been one of the key challenges in data mining. In this paper we attempt to help on this task by proposing two stochastic models to search for infection hot spots using social media trajectories. Our application scenario is a major infectious disease in Brazil and other tropical countries, dengue. We applied our models to data from 11 Brazilian cities and were able to detect infection hot spots in 4 of them. This result shows the usefulness of our methods to disease surveillance. To identify the high risk regions would be invaluable to direct preventive efforts and mitigation actions. Currently, we are carrying out a validation procedure of our results with local health officials.
We see our proposal as a first step on the direction of a more general and comprehensive framework. In fact, future research directions abound, both from theoretical and practical perspectives. One direction is to incorporate a richer data structure allowing features to be included at the individual level. In this paper, we only considered a binary indicator (case or control). However, we could add other features such as age and sex of the individuals. Another possibility is to associate features to the events that constitute the trajectories. For instance, distinguishing whether the event occurred in the summer or winter is potentially useful. A third possible direction is to consider the social links between the individuals as a means to create a social network between the trajectories. Notwithstanding these further developments, our models are useful for the difficult task of infection hot spots detection.
References
Twitter: The Streaming API. https://dev.twitter.com/streaming/overview
World Health Organization. http://www.who.int/csr/disease/dengue/denguenet
Bhatt, S., et al.: The global distribution and burden of dengue. Nature 496, 504–507 (2013)
Chen, F., Neill, D.B.: Non-parametric scan statistics for event detection and forecasting in heterogeneous social media graphs. In: Proceedings of the 20th ACM SIGKDD Conference, pp. 1166–1175 (2014)
Duczmal, L., Assunção, R.: A simulated annealing strategy for the detection of arbitrarily shaped spatial clusters. Comput. Stat. Data Anal. 45(2), 269–286 (2004)
Gomide, J., Veloso, A., Meira Jr., W., Almeida, V., Benevenuto, F., Ferraz, F., Teixeira, M.: Dengue surveillance based on a computational model of spatio-temporal locality of twitter. In: Proceedings of the ACM WebSci Conference (2011)
Kulldorff, M., Nagarwalla, N.: Spatial disease clusters: detection and inference. Stat. Med. 14(8), 799–810 (1995)
Kulldorff, M.: A spatial scan statistic. Comm. Stat. Theory Meth. 26(6), 1481–1496 (1997)
Kulldorff, M., Heffernan, R., Hartman, J., Assunção, R., Mostashari, F.: A space time permutation scan statistic for disease outbreak detection. PLoS Med (2005)
Lima, A., Stanojevic, R., Papagiannaki, D., Rodriguez, P., González, M.C.: Understanding individual routing behaviour. Roy. Soc. Interface 13(116) (2016)
McFowland III, E., Speakman, S., Neill, D.B.: Fast generalized subset scan for anomalous pattern detection. J. Mach. Learn. Res. 14, 1533–1561 (2013)
Murray, N.E.A., Quam, M.B., Wilder-Smith, A.: Epidemiology of dengue: past, present and future prospects. Clin. Epidemiol. 5, 299–309 (2013)
Neill, D.B., Cooper, G.F.: A multivariate Bayesian scan statistic for early event detection and characterization. Mach. Learn. 79(3), 261–282 (2010)
Neill, D.B., Moore, A.W.: Rapid detection of significant spatial clusters. In: Proceedings of the 10th ACM SIGKDD, pp. 256–265 (2004)
Neill, D.B., Moore, A.W., Sabhnani, M., Daniel, K.: Detection of emerging space-time clusters. In: Proceedings of the 11th ACM SIGKDD Conference, pp. 218–227 (2005)
Sakaki, T., Okazaki, M., Matsuo, Y.: Earthquake shakes twitter users: real-time event detection by social sensors. In: Proceedings of WWW, pp. 851–860 (2010)
Shi, L., Janeja, V.P.: Anomalous window discovery through scan statistics for linear intersecting paths (SSLIP). In: Proceedings of the 15th SIGKDD, pp. 767–776 (2009)
Silva, T.H., de Melo, P.O.V., Almeida, J., Loureiro, A.A.: Large-scale study of city dynamics and urban social behavior using participatory sensing. IEEE Wirel. Commun. 21(1), 42–51 (2014)
Souza, R.C.S.N.P., de Brito, D.E.F., Cardoso, R.L., de Oliveira, D.M., Meira Jr., W., Pappa, G.L.: An evolutionary methodology for handling data scarcity and noise in monitoring real events from social media data. In: Bazzan, A.L.C., Pichara, K. (eds.) IBERAMIA 2014. LNCS, vol. 8864, pp. 295–306. Springer, Heidelberg (2014)
Veloso, A., Meira Jr., W., Zaki, M.J.: Lazy associative classification. In: Proceedings of the International Conference on Data Mining, pp. 645–654 (2006)
Wu, M., Song, X., Jermaine, C., Ranka, S., Gums, J.: A LRT framework for fast spatial anomaly detection. In: 15th ACM SIGKDD, pp. 887–896 (2009)
Yoo, J.S., Bow, M.: Mining spatial colocation patterns: a different framework. Data Min. Knowl. Disc. 24, 159–194 (2012)
Zheng, Y.: Trajectory data mining: an overview. ACM Trans. Intell. Syst. Technol. 6 (2015)
Zhou, R., Shu, L., Su, Y.: An adaptive minimum spanning tree test for detecting irregularly-shaped spatial clusters. Comput. Stat. Data Anal. 89, 134–146 (2015)
Acknowledgements
This work was partially funded by Fapemig, CNPq, CAPES, and by the projects MASWeb (FAPEMIG-PRONEX APQ-01400-14), InWeb (MCT/CNPq 573871/2008-6) and EUBra-BIGSEA (H2020-EU.2.1.1 690116, Brazil/MCTI/RNP GA-000650/04).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Souza, R.C.S.N.P., Assunção, R.M., de Oliveira, D.M., de Brito, D.E.F., Meira, W. (2016). Infection Hot Spot Mining from Social Media Trajectories. In: Frasconi, P., Landwehr, N., Manco, G., Vreeken, J. (eds) Machine Learning and Knowledge Discovery in Databases. ECML PKDD 2016. Lecture Notes in Computer Science(), vol 9852. Springer, Cham. https://doi.org/10.1007/978-3-319-46227-1_46
Download citation
DOI: https://doi.org/10.1007/978-3-319-46227-1_46
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46226-4
Online ISBN: 978-3-319-46227-1
eBook Packages: Computer ScienceComputer Science (R0)