Abstract
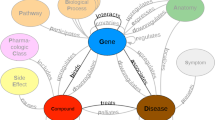
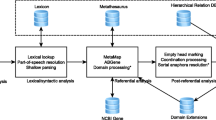
Identifying new potential treatment options (say, medications and procedures) for known medical conditions that cause human disease burden is a central task of biomedical research. Since all candidate drugs cannot be tested with animal and clinical trials, in vitro approaches are first attempted to identify promising candidates. Even before this step, due to recent advances, in silico or computational approaches are also being employed to identify viable treatment options. Generally, natural language processing (NLP) and machine learning are used to predict specific relations between any given pair of entities using the distant supervision approach. In this paper, we report preliminary results on predicting treatment relations between biomedical entities purely based on semantic patterns over biomedical knowledge graphs. As such, we refrain from explicitly using NLP, although the knowledge graphs themselves may be built from NLP extractions. Our intuition is fairly straightforward – entities that participate in a treatment relation may be connected using similar path patterns in biomedical knowledge graphs extracted from scientific literature. Using a dataset of treatment relation instances derived from the well known Unified Medical Language System (UMLS), we verify our intuition by employing graph path patterns from a well known knowledge graph as features in machine learned models. We achieve a high recall (92 %) but precision, however, decreases from 95 % to an acceptable 71 % as we go from uniform class distribution to a ten fold increase in negative instances. We also demonstrate models trained with patterns of length \(\le 3\) result in statistically significant gains in F-score over those trained with patterns of length \(\le 2\). Our results show the potential of exploiting knowledge graphs for relation extraction and we believe this is the first effort to employ graph patterns as features for identifying biomedical relations.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Although SemMedDB has 70 million relations, there are many duplicates given a relation can be extracted from multiple sentences due to the semantic mapping to UMLS concepts and semantic network predicates.
References
Kilicoglu, H., Shin, D., Fiszman, M., Rosemblat, G., Rindflesch, T.C.: SemMedDB: a pubmed-scale repository of biomedical semantic predications. Bioinformatics 28(23), 3158–3160 (2012)
Kim, S., Liu, H., Yeganova, L., Wilbur, W.J.: Extracting drug-drug interactions from literature using a rich feature-based linear kernel approach. J. Biomed. Inform. 55, 23–30 (2015)
Lu, Z.: PubMed and beyond: a survey of web tools for searching biomedical literature. Database J. Biol. Databases Curation (2011)
Mintz, M., Bills, S., Snow, R., Jurafsky, D.: Distant supervision for relation extraction without labeled data. In: Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP, pp. 1003–1011. Association for Computational Linguistics (2009)
National Library of Medicine. Current Hierarchy of UMLS Predicates. http://www.nlm.nih.gov/research/umls/META3_current_relations.html
National Library of Medicine. Current Hierarchy of UMLS Semantic Types. http://www.nlm.nih.gov/research/umls/META3_current_semantic_types.html
National Library of Medicine. Semantic MEDLINE Database.http://skr3.nlm.nih.gov/SemMedDB/
National Library of Medicine. SemRep - NLM’s Semantic Predication Extraction Program. http://semrep.nlm.nih.gov
National Library of Medicine. Unified Medical Language System Reference Manual. http://www.ncbi.nlm.nih.gov/books/NBK9676/
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P., Weiss, R., Dubourg, V., Vanderplas, J., Passos, A., Cournapeau, D., Brucher, M., Perrot, M., Duchesnay, E.: Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011)
Riedel, S., Yao, L., McCallum, A.: Modeling relations and their mentions without labeled text. In: Balcázar, J.L., Bonchi, F., Gionis, A., Sebag, M. (eds.) ECML PKDD 2010, Part III. LNCS, vol. 6323, pp. 148–163. Springer, Heidelberg (2010)
Rindflesch, T.C., Fiszman, M.: The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J. Biomed. Inform. 36(6), 462–477 (2003)
Ritter, A., Zettlemoyer, L., Etzioni, O., et al.: Modeling missing data in distant supervision for information extraction. Trans. Assoc. Comput. Linguist. 1, 367–378 (2013)
Surdeanu, M., Tibshirani, J., Nallapati, R., Manning, C.D.: Multi-instance multi-label learning for relation extraction. In: Proceedings of the 2012 Conference on Empirical Methods in Natural Language Processing, pp. 455–465. Association for Computational Linguistics (2012)
Xu, W., Hoffmann, R., Zhao, L., Grishman, R.: Filling knowledge base gaps for distant supervision of relation extraction. In: Proceedings of the 51st Annual Meeting of the Association for Computational Linguistics, pp. 665–670. Association for Computational Linguistics (2013)
Zhou, Z.-H.: Ensemble Methods: Foundations and Algorithms. CRC Press, Boca Raton (2012)
Acknowledgments
Thanks to anonymous reviewers for their helpful comments that helped improve the paper. The project described in this paper was supported by the National Center for Advancing Translational Sciences (UL1TR000117). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Bakal, G., Kavuluru, R. (2015). Predicting Treatment Relations with Semantic Patterns over Biomedical Knowledge Graphs. In: Prasath, R., Vuppala, A., Kathirvalavakumar, T. (eds) Mining Intelligence and Knowledge Exploration. MIKE 2015. Lecture Notes in Computer Science(), vol 9468. Springer, Cham. https://doi.org/10.1007/978-3-319-26832-3_55
Download citation
DOI: https://doi.org/10.1007/978-3-319-26832-3_55
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-26831-6
Online ISBN: 978-3-319-26832-3
eBook Packages: Computer ScienceComputer Science (R0)