Abstract
Segmentation based on active contour has been received widespread concerns recently for its good flexible performance. However, most available active contour models lack adaptive initial contour and priori information of target region. In this paper, we presented a new method that is based on active contours combined with saliency map for plant leaf segmentation. Firstly, priori shape information of target objects in input leaf image which is used to describe the initial curve adaptively is extracted with the visual saliency detection method in order to reduce the influence of initial contour position. Furthermore, the proposed active model can segment images adaptively and automatically. Experiments on two applications demonstrate that the proposed model can achieve a better segmentation result.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Currently, precision agriculture has become one of the frontier researches in the field of agriculture. Promote the use of precision agriculture could make use of resources reasonably, increasing crop yields, cut production costs and improve agricultural competitiveness. But the most relevant studies are based on the basic data of crop growth, which related with the shape features computation of plant leaf.
In agriculture, numerous image-processing based computerized tools have been developed to help farmers to monitor the proper growth of their crops. Special attention has been put towards the latest stages when crop is near harvesting. For example, at the time of harvesting, some computer tools are used to discriminate between plants and other objects present in the field. In the case of machines that uproot weeds, they have to discriminate between plants and weeds, whilst in the case of machines that harvest; they have to differentiate one crop from the other.
Previous studies have identified the challenges and have successfully produced systems that address them. The requirements for reduced production costs, the needs of organic agriculture and the proliferation of diseases have been the driving forces for improving the quality and quantity of food production. In fact, the main source of plant diseases comes from leaves, such as leaf spot, rice blast, leaf blight, etc. When plants become diseased, they can display a range of symptoms such as colored spots, or streaks that can occur on the leaves, stems, and seeds of the plant. These visual symptoms continuously change their color, shape and size as the disease progresses.
Thus, in the area of disease control, most researches have been focused on the treatment and control of weeds, but few studies focused on the automatic identification of diseases. Automatic plant disease identification by visual inspection can be of great benefit to those users who have little or no information about the crop they are growing. Such users include farmers, in underdeveloped countries, who cannot afford the services of an expert agronomist, and also those living in remotes areas where access to assistance via an internet connection can become a significant factor.
During the past few decades,there has been substantial progress in the field of image segmentation and its application, that’s a prerequisite for computer vision and field management automation. Camargo [1] study an image-processing based algorithm to automatically identify plant disease visual symptoms, the processing algorithm developed starts by converting the RGB image of the diseased plant or leaf, into the H, I3a and I3b color transformations, then segmented by analyzing the distribution of intensities in a histogram. Minervini [2] proposed an image-based plant phenotyping with incremental learning and active contours for accurate plant segmentation. This method is a vector valued level set formulation that incorporates features of color intensity, local texture, and prior knowledge. Sonal [3] presented a classification algorithm of cotton leaf spot disease using support vector machine. Prasad [4] explores a new dimension of pattern recognition to detect crop diseases based on Gabor Wavelet Transform. But there are many difficulties during leaf image process such as complex color components, irregular texture distribution, light shadow, and other random noises, even with some of complex and heterogeneity background, so leaf segmentation is still a nut for image analysis.
Recently, segmentation algorithms based on active contours has been received widespread concerns by many researchers due to their variable forms, flexible structure and excellent performance. However, most available active contour models suffer from lacking adaptive initial contour and priori information of target region. In this paper, we presented a new object segmentation method that is based on active contours with combined saliency map. Firstly, priori shape information of target objects in input images which is used to describe the initial curve adaptively is extracted with the visual saliency detection method in order to reduce the influence of initial contour position. Furthermore, the proposed active model can segment images adaptively and automatically, and the segmented results accord with the property of human visual perception. Experimental results demonstrate that the proposed model can achieve better segmentation results than some traditional active contour models. Meanwhile our model requires less iteration and with better computation efficiency than traditional active contours methods although the background of the image is clutter.
2 Saliency Detect Model
When it comes to the concept of saliency, another term visual attention is often referred. While the terms attention, saliency are often used interchangeably, each has a more subtle definition that allows their delineation. Attention is a general concept covering all factors that influence selection mechanisms, whether they are scene-driven bottom-up or expectation-driven top-down. Saliency intuitively characterizes some parts of a scene-which could be objects or regions-that appear to an observer to stand out relative to their neighboring parts. The term “salient” is often considered in the context of bottom-up computations [5]. Modeling visual attention—particularly stimulus-driven, saliency-based attention—has been a very active research area over the past 25 years. Many different models of attention are now available, which aside from lending theoretical contributions to other fields, have demonstrated successful applications in computer vision, mobile robotics, and cognitive systems.
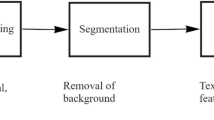
Our saliency detect algorithm consists of three parts (Fig. 1):
Firstly, color-complexity features extraction. Decomposes a given image into a color histogram after quantizing each color channel to have 12 different values to speed up our algorithm, which once proposed by Cheng et al. [6]. And then compute color contrast differences between colors over a global image to construct a saliency map.
Secondly, color-spatial features extraction. In order to establish the color distributions of pixels in an image, k-means is exploited to find k centers of masses for each color. And a relative minimum method was proposed to calculate spatial distances between different colors to obtain the spatial weightings. Then saliency map was adjusted by spatial weightings.
Lastly, a smoothing procedure (see also [6]) is used to reduce the noise disturbance due to quantization.
Overall, our method integrated the contrast map with colors spatial distribution to derive a saliency measure that produces a pixel-accurate saliency map which uniformly covers the region of interest and consistently separates foreground and background. Specifically details can refer to our paper in [7].
2.1 Color-Complexity Feature
A saliency value of a pixel is definition by its differences with all other pixels in image. Formula (1) is the pixel I i saliency value in an image I,
where CD(I i , I j ) is the color distance metric between pixels I i and I j in the L*a*b* color space [6, 8]. The following formula (2) is obtained by expanding this equation,
where N is the number of pixels in image I. Its means that pixel with the same color value would be have a same saliency value according to (2). Thus (2) need further restructured, such that the terms with the same color c q are rearranged to be together,
where c p represents the color of pixel I i , f q is the occurrence frequency of color c q in image I and n is the total number of colors Color category. The histogram was exploited to make a statistics of color distribution for its simple and rapid process.
The computational complexity of Eq. (3) is O(N) + O(n 2), which means that reducing the number of colors n is a key for computation speed up. The color has 2563 possible values, where the value of each color channel locates in the range of [0, 255]. Zhai and Shah [8] reduce the number of colors to n 2 = 2562 only adopted luminance, because of its only employs two color channels, whereas the information of the third color channel was ignored. In this paper, full color space instead of luminance was used by dividing each color channel into 12 values for reduce the number of colors to n 3 = 123 rapidly, which once proposed by Cheng et al. [6] At the same time, considering that some colors just appears rarely and the resulting saliency map almost suffered from no affect if without them, so we can further reduce n by ignoring those colors that with less occurring frequencies. Thus by this means, the number of colors n was reduced greatly which made computation more efficiently.
2.2 Color-Spatial Feature
Performance comparison of HC and RC [6] implies that taking spatial information into account could improve the results substantially. To the same contrast difference extent, those surrounding regions have more contribution for a region saliency than those far-away regions. So naturally we integrate spatial weighting to increase the effects of closer regions and decrease the effects of farther regions.
According to the foregoing discussion, the number of colors n was reduced to a considerable small number around by choosing more frequently occurring colors and ensuring these colors cover the colors of more than 95 % of the image pixels, we typically are left with around n = 85 colors. Then calculate the spatial distance between colors after finding k mass centers exploiting k-means for each color. The initial value k for k-means algorithm should be decided by taking both accuracy and efficiency of saliency computation into consideration.
Figure 2 shows details of how spatial distance computed between colors c p and c q . For illustrates convenience here k = 3. In Fig. 2, color c p has three mass centers, represented by yellow points, from top to bottom in turn is c p,1, c p,2, c p,3. Likewise, green points (i.e. c q,1, c q,2, c q,3) are the mass centers of color c q . We use set p and set q to represent the mass centers of colors c p and c q respectively for illustrate clearly. Then the algorithm of compute the relative minimum distance between these two sets as follow:
-
(1)
For c p,1 in set p , find the closest point to it in set q , in this example below is c q,1. Then they form pairwise naturally.
-
(2)
Accordingly, c p,1 and c q,1 are removed from its set respectively.
-
(3)
Continually for the remaining points in set p likes step 1 and 2, until set p is empty.
The spatial distance of the two colors is defined in (4).
where k is the number of cluster centers, || . || represent the point distance metric. Then the saliency value for each color is defined as,
where c p is the color of pixel I i , n denotes the number of colors, CD(c p , c q ) is the color distance metric between c p and c q in L*a*b color space. And
is the spatial weighting between colors c p and c q , where σ s controls the strength of spatial weighting.
Finally, a smoothing procedure will be taken to refine the saliency value for each color. The saliency value of each color is adjusted to the weighted average of the saliency values of similar colors,
where m is the number of nearest colors to c, and we choose m = n/4, \( M = \sum\nolimits_{i = 1}^{m} {CD(c,c_{i} )} \) is the sum of distances between color c and its m nearest neighbors c i .
3 Salient Region-Based Active Contour Model
While the above steps provide a collection of regions of saliency and an initial rough segmentation, the goal of this section is to obtain a highly accurate segmentation of each plant. Operating on a smaller portion of the image allows us to use more complex algorithms, which likely would not have been efficient and effective in the full image. The motivation for using an active contour method for object segmentation is its ability to model arbitrarily complex shapes and handle implicitly topological changes. Thus, the level set based segmentation method can be effectively used for extracting a foreground layer with fragmented appearance, such as leaves of the same plant.
Let Ω ⊂ ℜ2 is the 2D image space, and I: Ω → ℜ be a given gray level image. In [9], Mumford and Shah formulated the image segmentation problem as follows: given an image I, finding a contour C which segments the image into non-overlapping regions. The energy functional was proposed as following:
Where |C| is the contour length, μ, v > 0 are constants to balance the terms. The minimization of Mumford–Shah functional results in an optimal contour C that segments the given image I, u is an image to approximate the original image I, which is smooth within each region inside or outside the contour C. In practice, it is difficult to minimize the functional (8) due to the unknown contour C of lower dimension and the non-convexity of the functional.
To overcome the difficulties in solving Eq. (8), Chan and Vese [10] presented an active contour model to the Mumford–Shah problem for a special case where the image u in the functional (8) is a piecewise constant function. They proposed to minimize the following energy functional:
Where outside (C) and inside (C) represent the regions outside and inside the contour C, respectively, and c 1 and c 2 are two constants that approximate the image intensity in outside (C) and inside (C).
But this Chan-Vese model does not suitable for our plant leaf processing, because the model has some difficulties for vector-valued color image with intensity inhomogeneity. Then Chan-Sandberg-Vese proposed a new algorithm in [11] for object detection in vector-valued images (such as RGB or multispectral). In this paper, since we want to take advantage of the existence of multiple image features (in the following we refer to them as channels) we build our salient region-based active contour model F SRAC for vector valued images. Suppose we obtained an initial contour C that is a shape approximation of the object boundary through the saliency detection method mentioned above, and the image I divided into two parts-salient region Ω S and non salient region Ω NS by the contour C. Then add this prior shape knowledge into F SRAC, and the overall energy functional for F SRAC following the formulation in [11] is defined as:
where z denotes a pixel location in an image channel \( I_{i} ,i = 1\ldots N,\lambda_{i}^{ + } \) and \( \lambda_{i}^{ - } \) define the weight of each term (inside and outside the contour), \( \overrightarrow {c}^{ - } \) is the vector valued representation of the mean for each channel outside the contour, and \( \overrightarrow {c}^{ + } \) and \( \overrightarrow {m}^{ + } \) are the vector valued representations of the mean and median respectively for each channel inside the contour. The way we estimate these statistical quantities will be described shortly.
Following standard level set formulations [11], we replace the contour curve C with the level set function ϕ [12]: \( F^{SRAC} (\phi ,\overrightarrow {c}^{ + } ,\overrightarrow {m}^{ + } ,\overrightarrow {c}^{ - } ) \).
The vectors \( \overrightarrow {c}^{ + } \), \( \overrightarrow {m}^{ + } \) and \( \overrightarrow {c}^{ - } \) are defined in similar fashion to other intensity driven active contour models as statistical averages and medians:
for each channel I i , i = 1…N, inside or outside the contour.
Using the level set function ϕ to represent the contour C in the image domain Ω, the energy functional can be written as follows:
where H is the Heaviside function.
By keeping \( \overrightarrow {c}^{ + } \), \( \overrightarrow {m}^{ + } \) and \( \overrightarrow {c}^{ - } \) are fixed, we minimize the energy function \( F^{SRAC} (\phi ,\overrightarrow {c}^{ + } ,\overrightarrow {m}^{ + } ,\overrightarrow {c}^{ - } ) \) with respect to ϕ to obtain the gradient descent flow as:
where δ is the Dirac delta function.
4 Application and Experimental
4.1 Image-Based Plant Phenotyping Detect
Understanding biological function and the complex processes involved in the development of plants relies on understanding the interaction between genetic information and the environment, and how they affect the phenotype (the appearance or behavior) of the organism and consequently desirable traits. Model plant systems, such as Arabidopsis thaliana, have been used extensively for this purpose. However, as of today, inexpensive and automated phenotyping (phenomics) remains a bottleneck. Until recently most phenotypes (e.g., related to plant growth) were acquired in destructive ways (e.g., weigh the plant, or cut out and measure a leaf) or involved human survey (e.g., measuring leaf size or plant radius) in situ without destructing the plant. Naturally these methods are faced with low throughput and high productivity cost. Consequently, there has been a growing interest towards developing solutions for the automated analysis of visually observable traits of the plants [2].
In this section we propose a method adopt model mentioned above for the automated segmentation and analysis of plant images from phenotyping experiments of Arabidopsis rosettes. We use the image dataset in [13] which acquired in a general laboratory setting with a static camera that captures many plants at the same time for this plant segmentation test.
Prior Knowledge Map: From a computer vision perspective, a laboratory setup for plant phenotyping experiments presents several challenges such as neon light illumination, water reflection, shadows, and moss, contributing to noise and scene complexity. To eliminate issues of non uniform illumination (due to lighting distortion from neon lights and shadowing), when utilizing color information we convert the RGB color space to ExG = 2G-R-B, where R, G, B are the three components of RGB space, because the most colors of plant leaf in this test are green.
Map Fusion: After got the saliency map and the prior knowledge map, then we fusing them again through the following formula get the final map:
where Map f is fusing map, Map s and Map p denotes saliency map and prior map respectively, but Map p must be tuned with same format as Map s and normalized.
Finally, after the fusion map is generated by the formula (14), it is then binarized segmented by an adaptive threshold T a proposed in [14], defined as twice the mean saliency of the Map f image:
where W and H are the width and the height of the saliency map respectively.
And the contour of the thresholded response forms the initial curve for the active contour model. The maximum number of iterations for contour evolution is set to 300, and we used \( \lambda_{i}^{ + } = \lambda_{i}^{ - } = 1 \), for all i.
Post-processing: After the segmentation is performed based on the active contour model, small blobs containing foreground pixels are removed in a way similar to the procedure used in [15]. To be specific, all holes within each blob are filled by morphological operations and small blobs are removed based on the number of pixels in each filled blob. Empirically, a small blob is determined by the following criterion, that is, one smaller than the half of the largest blob: 0.5 × max b∈B |b| where b is an individual blob, B is the set of blobs and | · | the number of pixels in the blob.
To compare the segmentation performance between the “reference segmentation” marked by specialists in [13] and an outline from our model. We employed the following metrics:
where TP, FP, and FN represent the number of true positive, false positive, and false negative pixels, respectively, calculated by comparing algorithmic result and ground-truth segmentation masks. Precision is the fraction of pixels in the segmentation mask that matches the ground truth, whereas recall is the fraction of ground-truth pixels contained in the segmentation mask. The Dice similarity coefficients are used to measure the spatial overlap between algorithmic result and ground truth. All of these metrics are expressed in percentages, with larger values representing higher agreement between ground truth and algorithmic result.
This level of accuracy is observed across in [13] dataset. Table 1 reports averaged results of segmentation accuracy over the whole dataset. The Reference method is an approach based on K-means, due to its widespread adoption in plant segmentation which shows poor accuracy in terms of precision and Dice, and a very high recall value due to the constant over-segmentation (i.e., the plant is fully contained in the segmentation mask, along with large portions of earth and moss from the background). The method in [2] appears the best in results for its balance in all influence factors. Our proposed system also achieves a good accuracy in this test as show in Table 1 and Fig. 3.
4.2 Diseased Plant Leaf Detect
This section describes an image processing based our method for identifies the visual symptoms of plant diseases, from an analysis of colored images. The plant diseases model is basically the same as the 4.1 above. Whereas the prior knowledge map in here is not the ExG but an R-channel in RGB map because the most color of plant diseases location is a deep color such as yellow-red. The adaptive threshold T a defined as 1.5 times of the mean saliency Map f here. The test image dataset we used was referred to [1], which include 20 diseased leaves images. The experimental comparison illustrated as follow examples in Fig. 4 and Table 2 demonstrates that our model has a nearly good segmentation effect as [1], but we provided a novelty approach.
5 Conclusions
In this paper, we present a new plant leaf image segmentation method that is based on active contours combined with saliency map. It is known that saliency region detect can easily attain the approximate contour of the desirable object in image, and then set it as the initial position of evolution curve for active contour model to construct a new level set energy functional with a more faster evolution speed and more accurate object segmentation. Finally, experiments comparison on two applications demonstrates that our proposed model has a good effect on plant leaf image segmentation.
References
Camargo, A., Smith, J.S.: An image-processing based algorithm to automatically identify plant disease visual symptoms. Biosyst. Eng. 102(1), 9–21 (2009)
Minervini, M., Abdelsamea, M.M., Tsaftaris, S.A.: Image-based plant phenotyping with incremental learning and active contours. Ecol. Inform. 23, 35–48 (2014)
Patil, S.P.: Classification of cotton leaf spot disease using support vector machine. J. Eng. Res. Appl. 4, 92–97 (2014)
Prasad, S., Kumar, P., Hazra, R., Kumar, A.: Plant leaf disease detection using Gabor wavelet transform. In: Panigrahi, B.K., Das, S., Suganthan, P.N., Nanda, P.K. (eds.) SEMCCO 2012. LNCS, vol. 7677, pp. 372–379. Springer, Heidelberg (2012)
Borji, A., Itti, L.: State-of-the-art in visual attention modeling. IEEE Trans. Pattern Anal. Mach. Intell. 35(1), 185–207 (2013)
Cheng, M.M., Zhang, G.X., Mitra, N.J., Huang, X.L., Hu, S.M.: Global contrast based salient region detection. In: CVPR, pp. 409–416 (2011)
Wang, Z., Li, L., Chen, Y., et al.: Salient region detection based on color-complexity and color-spatial features. In: 2013 Fourth International Conference on Intelligent Control and Information Processing (ICICIP), pp. 699–704. IEEE (2013)
Zhai, Y., Shah, M.: Visual attention detection in video sequences using spatiotemporal cues. In: ACM Multimedia, pp. 815–824 (2006)
Mumford, D., Shah, J.: Optimal approximations by piecewise smooth functions and associated variational problems. Commun. Pure Appl. Math. 42(5), 577–685 (1989)
Chan, T.F., Vese, L.A.: Active contours without edges. IEEE Trans. Image Process. 10(2), 266–277 (2001)
Chan, T.F., Sandberg, B.Y., Vese, L.A.: Active contours without edges for vector-valued images. J. Vis. Commun. Image Represent. 11(2), 130–141 (2000)
Zhao, H.K., Chan, T., Merriman, B., et al.: A variational level set approach to multiphase motion. J. Comput. Phys. 127(1), 179–195 (1996)
Achanta, R., Hemami, S., Estrada, F., Süsstrunk, S.: Frequency-tuned salient region detection. In: CVPR, pp. 1597–1604 (2009)
Koh, J., Kim, T., Chaudhary, V., Dhillon, G.: Segmentation of the spinal cord and the dural sac in lumbar mr images using gradient vector flow field. In: EMBC, pp. 3117–3120 (2010)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Qiangqiang, Z., Zhicheng, W., Weidong, Z., Yufei, C. (2015). Contour-Based Plant Leaf Image Segmentation Using Visual Saliency. In: Zhang, YJ. (eds) Image and Graphics. ICIG 2015. Lecture Notes in Computer Science(), vol 9218. Springer, Cham. https://doi.org/10.1007/978-3-319-21963-9_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-21963-9_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-21962-2
Online ISBN: 978-3-319-21963-9
eBook Packages: Computer ScienceComputer Science (R0)