Abstract
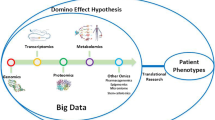
To understand the mechanism of biological processes inside a human, it is necessary to look at its various regulatory aspects, such as DNA methylation and post-translational modifications of histones (PTMs). These characteristics are all susceptible to disease-induced alterations in cell signalling and phenotypes. We need to use a multi-omics approach because many illnesses result from complex processes, and we must examine each of these traits and their interactions to gain insights into the causes of diseases. Therefore, investigating multi-omics data is a crucial aspect of molecular-level healthcare research and has yielded cutting-edge discoveries. High-throughput technologies are becoming more widely available, which has led to an increase in the amount of omics data being produced. These omics data include epigenomics, transcriptomics and genomics, proteomics which all aim to represent various but complementary biological layers. By making it possible to thoroughly examine biological systems and molecular underpinnings of disease development, these data have changed healthcare research. There is a strong trend toward adding multi-omics analysis into healthcare research to explain the intricate interactions across molecular levels, even if the integration and translation of multi-omics data into relevant functional insights remains a significant barrier. Multi-omics data can help improve prevention, early detection, and prediction, monitor history, interpret patterns and design a personalised treatment. Various Machine Learning algorithms grouped under supervised and unsupervised learning techniques have been used to integrate data through various omics levels. This multi-omics analysis has various applications in deciphering the causative reason for many diseases like cancer and thus has helped in taking a step forward towards personalised medicine for tailoring the right medication for the right person. Hence, a lot of attention is given to establishing various machine learning algorithms for the automatic integration of multi-omics data. With this data, machine learning algorithms can be employed to produce diagnostic and classification biomarkers, offering fresh information. However, researchers have identified a bulk of biomarkers that consider only one omics parameter at a time and have not properly utilised a recent multi-omics research strategy, which can adequately capture the complexity of biological systems. The complementary knowledge that each omics layer contributes must be included in multi-omics data integration strategies. As a result, it is advisable to support the development of novel machine—learning methods. This chapter outlines the roadmap for multi-omics integration with machine learning, various integration methods, challenges, and future aspects.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Graw, S., Chappell, K., Washam, C.L., Gies, A., Bird, J., Robeson, M.S., Byrum, S.D.: Multi-omics data integration considerations and study design for biological systems and disease. Mol. Omics 17(2), 170–185 (2021). https://doi.org/10.1039/D0MO00041H
Santiago-Rodriguez, T.M., Emily, B.: Multi ‘omic data integration: a review of concepts, considerations, and approaches. In: Seminars in Perinatology, p. 151456. WB Saunders (2021). https://doi.org/10.1016/j.semperi.2021.151456
Picard, M., Scott-Boyer, M.P., Bodein, A., Périn, O., Droit, A.: Integration strategies of multi-omics data for machine learning analysis. Comput. Struct. Biotechnol. J. 19, 3735–3746 (2021). https://doi.org/10.1016/j.csbj.2021.06.030
Subramanian, I., Verma, S., Kumar, S., Jere, A., Anamika, K.: Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights 14, 1177932219899051 (2020). https://doi.org/10.1177/1177932219899051
Huang, S., Chaudhary, K., Garmire, L.X.: More is better: recent progress in multi-omics data integration methods. Front. Genet. 8, 84 (2017). https://doi.org/10.3389/fgene.2017.00084
Reel, P.S., Reel, S., Pearson, E., Trucco, E., Jefferson, E.: Using machine learning approaches for multi-omics data analysis: a review. Biotechnol. Adv. 49, 107739 (2021). https://doi.org/10.1016/j.biotechadv.2021.107739
Cai, Z., Poulos, R.C., Liu, J., Zhong, Q.: Machine learning for multi-omics data integration in cancer. iScience 22, 103798 (2022). https://doi.org/10.1016/j.isci.2022.103798
Bansal, H., Luthra, H., Chaurasia, A.: Impact of machine learning practices on biomedical informatics, its challenges and future benefits. In: Artificial Intelligence Technologies for Computational Biology, pp. 273–294. CRC Press (2023). https://doi.org/10.1201/9781003246688-12
Arjmand, B., Hamidpour, S.K., Tayanloo-Beik, A., Goodarzi, P., Aghayan, H.R., Adibi, H., Larijani, B.: Machine learning: a new prospect in multi-omics data analysis of cancer. Front. Genet. 13, 76 (2022). https://doi.org/10.3389/fgene.2022.824451
El-Manzalawy, Y., Hsieh, T.Y., Shivakumar, M., Kim, D., Honavar, V.: Min-redundancy and max-relevance multi-view feature selection for predicting ovarian cancer survival using multi-omics data. BMC Med. Genomics 11(3), 19–31 (2018). https://doi.org/10.1186/s12920-018-0388-0
Wang, B., Mezlini, A.M., Demir, F., Fiume, M., Tu, Z., Brudno, M., Haibe-Kains, B.: Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 11(3), 333–337 (2014). https://doi.org/10.1038/nmeth.2810
Lan, L., Djuric, N., Guo, Y., Vucetic, S.: MS-k NN: protein function prediction by integrating multiple data sources. BMC Bioinform. 14(Suppl 3), S8 (2013). https://doi.org/10.1186/1471-2105-14-S3-S8
Quinlan, J.R.: Induction of decision trees. Mach. Learn. 1, 81–106 (1986). https://doi.org/10.1007/BF00116251
Gligorijević, V., Pržulj, N.: Methods for biological data integration: perspectives and challenges. J. R. Soc. Interface 12(112), 20150571 (2015). https://doi.org/10.1098/rsif.2015.0571
Huang, S., Cai, N., Pacheco, P.P., Narrandes, S., Wang, Y., Xu, W.: Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics 15(1), 41–51 (2018). https://doi.org/10.21873/cgp.20063
Fawagreh, K., Gaber, M.M., Elyan, E.: Random forests: from early developments to recent advancements. Syst. Sci. Control Eng.: Open Access J. 2(1), 602–609 (2014). https://doi.org/10.1080/21642583.2014.956265
Shen, R., Olshen, A.B., Ladanyi, M.: Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics 25(22), 2906–2912 (2009). https://doi.org/10.1093/bioinformatics/btp543
Curtis, C., Shah, S., Chin, S.F., et al.: The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486(7403), 346–352 (2012). https://doi.org/10.1038/nature10983
Lock, E.F., Hoadley, K.A., Marron, J.S., Nobel, A.B., et al.: Joint and individual variation explained (JIVE) for integrated analysis of multiple data types. Ann. Appl. Stat. 7(1), 523 (2013). https://doi.org/10.1214/12-AOAS597
Hasin, Y., Seldin, M., Lusis, A.: Multi-omics approaches to disease. Genome Biol. 18(1), 1–15 (2017). https://doi.org/10.1186/s13059-017-1215-1
Xu, J., Wu, P., Chen, Y., Meng, Q., Dawood, H., Dawood, H.: A hierarchical integration deep flexible neural forest framework for cancer subtype classification by integrating multi-omics data. BMC Bioinform. 20(1), 1–11 (2019). https://doi.org/10.1186/s12859-019-3116-7
Bonnet, E., Calzone, L., Michoel, T.: Integrative multi-omics module network inference with Lemon-Tree. PLoS Comput. Biol. 11(2), e1003983 (2015). https://doi.org/10.1371/journal.pcbi.1003983
Yang, Y., Dong, X., Xie, B., Ding, N., Chen, J., Li, Y., Zhang, Q., Qu, H., Fang, X.: Databases and web tools for cancer genomics study. Genomics Proteomics Bioinform. 13(1), 46–50 (2015). https://doi.org/10.1016/j.gpb.2015.01.005
Tepeli, Y.I., Ünal, A.B., Akdemir, F.M., Tastan, O.: PAMOGK: a pathway graph kernel based multi-omics approach for patient clustering. Ph.D. Thesis. (2020)
Rappoport, N., Shamir, R.: NEMO: cancer subtyping by integration of partial multi-omic data. Bioinformatics 35(18), 3348–3356 (2019). https://doi.org/10.1093/bioinformatics/btz058
Reel, P.S., Reel, S., Pearson, E., Trucco, E., Jefferson, E.: Using machine learning approaches for multi-omics data analysis: a review. Biotechnol. Adv. 49, 107739 (2021). https://doi.org/10.1016/j.biotechadv.2021.107739
Chappell, K., Manna, K., Washam, C.L., Graw, S., Alkam, D., Thompson, M.D., Zafar, M.K., Hazeslip, L., Randolph, C., Gies, A., Bird, J.T.: Multi-omics data integration reveals correlated regulatory features of triple negative breast cancer. Mol. Omics 17(5), 677–691 (2021). https://doi.org/10.1039/d1mo00117e
Zhang, L., Lv, C., Jin, Y., Cheng, G., Fu, Y., Yuan, D., Tao, Y., Guo, Y., Ni, X., Shi, T.: Deep learning-based multi-omics data integration reveals two prognostic subtypes in high-risk neuroblastoma. Front. Genet. 9, 477 (2018). https://doi.org/10.3389/fgene.2018.00477
Kamburov, A., Cavill, R., Ebbels, T.M., Herwig, R., Keun, H.C.: Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics 27(20), 2917–2918 (2011). https://doi.org/10.1093/bioinformatics/btr499
Rohart, F., Gautier, B., Singh, A., Lê Cao, K.A.: mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13(11), e1005752 (2017). https://doi.org/10.1371/journal.pcbi.1005752
Meng, C., Kuster, B., Culhane, A.C., Gholami, A.M.: A multivariate approach to the integration of multi-omics datasets. BMC Bioinform. 15, 1–13 (2014). https://doi.org/10.1186/1471-2105-15-162
Argelaguet, R., Velten, B., Arnol, D., Dietrich, S., Zenz, T., Marioni, J.C., Buettner, F., Huber, W., Stegle, O.: Multi‐omics factor analysis—a framework for unsupervised integration of multi‐omics data sets. Mol. Syst. Biol. 14(6), e8124 (2018). https://doi.org/10.15252/msb.20178124
Bauer C., Stec, K., Glintschert, A., Gruden, K., Schichor, C., Or-Guil, M., Selbig, J., Schuchhardt, J.: BioMiner: paving the way for personalized medicine. Cancer Inform. 14, CIN. S20910 (2015). https://doi.org/10.4137/CIN.S20910
Tomczak, K., Czerwińska, P., Wiznerowicz, M.: Review the cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol./Współczesna Onkol. 2015(1), 68–77 (2015). https://doi.org/10.5114/wo.2014.47136
Wu, P., Heins, Z.J., Muller, J.T., Katsnelson, L., de Bruijn, I., Abeshouse, A.A., Schultz, N., Fenyö, D., Gao, J.: Integration and analysis of CPTAC proteomics data in the context of cancer genomics in the cBioPortal*[S]. Mol. Cell. Proteomics 18(9), 1893–1898 (2019). https://doi.org/10.1074/mcp.TIR119.001673
Shimada, K., Bachman, J.A., Muhlich, J.L., Mitchison, T.J.: shinyDepMap, a tool to identify targetable cancer genes and their functional connections from Cancer Dependency Map data. Elife 10, e57116 (2021). https://doi.org/10.7554/eLife.57116
García-Alcalde, F., García-López, F., Dopazo, J., Conesa, A.: Paintomics: a web-based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics 27(1), 137–139 (2011). https://doi.org/10.1093/bioinformatics/btq594
Misra, B.B., Langefeld, C., Olivier, M., Cox, L.A.: Integrated omics: tools, advances and future approaches. J. Mol. Endocrinol. 62(1), R21–R45 (2019). https://doi.org/10.1530/JME-18-0055
Subramanian, I., Verma, S., Kumar, S., Jere, A., Anamika, K.: Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights 14, 1177932219899051 (2020). https://doi.org/10.1177/1177932219899051
Luthra, H., Nihith, T.A.S., Pravallika, V.S.S., Raghuram Shree, R., Chaurasia, A., Bansal, H.: New paradigm in healthcare industry using big data analytics. In: IOP Conference Series: Materials Science and Engineering, p. 012054. IOP Publishing (2021). https://doi.org/10.1088/1757-899X/1099/1/012054
Bhattacharjya, R., Tiwari, A., Marella, T.K., Bansal, H., Srivastava, S.: New paradigm in diatom omics and genetic manipulation. Bioresour. Technol. 325, 124708 (2021). https://doi.org/10.1016/j.biortech.2021.124708
Bansal, H., Kohli, R.K., Saluja, K., Chaurasia, A.: Recent advancements in biomedical research in the era of AI and ML. Artif. Intell. Comput. Dyn. Biomed. Res. 8, 1–20 (2022). https://doi.org/10.1515/9783110762044-001
García, V., Sánchez, J.S., Marqués, A.I., Florencia, R., Rivera, G.: Understanding the apparent superiority of over-sampling through an analysis of local information for class-imbalanced data. Expert. Syst. Appl. 158 (2020). https://doi.org/10.1016/j.eswa.2019.113026
Bolívar, A., García, V., Florencia, R., Alejo, R., Rivera, G., Sánchez-Solís, J.P.: A preliminary study of smote on imbalanced big datasets when dealing with sparse and dense high dimensionality. In: Pattern Recognition: 14th Mexican Conference, MCPR 2022, Ciudad Juárez, Mexico, June 22–25, 2022, Proceedings, pp. 46–55. Springer International Publishing, Cham (2022). https://doi.org/10.1007/978-3-031-07750-0_5
Rivera, R., Florencia, R., García, V., Ruiz, A., Sánchez-Solís, J.P.: News classification for identifying traffic incident points in a Spanish-speaking country: a real-world case study of class imbalance learning. Appl. Sci. (Switzerland) 10(18) (2020). https://doi.org/10.3390/APP10186253
Leng, D., Zheng, L., Wen, Y., Zhang, Y., Wu, L., Wang, J., Wang, M., Zhang, Z., He, S., Bo, X.: A benchmark study of deep learning-based multi-omics data fusion methods for cancer. Genome Biol. 23(1), 1–32 (2022). https://doi.org/10.1186/s13059-022-02739-2
Nicora, G., Vitali, F., Dagliati, A., Geifman, N., Bellazzi, R.: Integrated multi-omics analyses in oncology: a review of machine learning methods and tools. Front. Oncol. 10, 1030 (2020). https://doi.org/10.3389/fonc.2020.01030
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Bansal, H., Luthra, H., Raghuram, S.R. (2023). A Review on Machine Learning Aided Multi-omics Data Integration Techniques for Healthcare. In: Rivera, G., Cruz-Reyes, L., Dorronsoro, B., Rosete, A. (eds) Data Analytics and Computational Intelligence: Novel Models, Algorithms and Applications. Studies in Big Data, vol 132. Springer, Cham. https://doi.org/10.1007/978-3-031-38325-0_10
Download citation
DOI: https://doi.org/10.1007/978-3-031-38325-0_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-38324-3
Online ISBN: 978-3-031-38325-0
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)