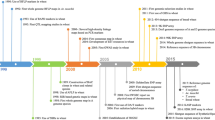
Abstract
Wheat is a crucial crop globally, with widespread cultivation and significant economic importance. To ensure food security amidst the increasing human population and new production challenges, such as climate change, it is imperative to develop novel wheat varieties that exhibit better quality, higher yield, and enhanced resistance to biotic and abiotic stress. To achieve this, leveraging comprehensive genomic resources from global breeding programs can aid in identifying within-species allelic diversity and selecting optimal allele combinations for superior cultivars. While previous single-reference genome assemblies have facilitated gene discovery and whole-genome level genotype–phenotype relationship modeling, recent research on variations within the pan-genome of all individuals in a plant species underscores their significance for crop breeding. We summarize the different approaches and techniques used for sequencing the large and intricate wheat genome, while highlighting the challenge of generating high-quality reference assemblies. We discuss the computational methods for building the pan-genome and research efforts that are aimed at utilizing the wheat pan-genome in wheat breeding programs.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
14.1 Introduction
In the early 2000s, technological advances in DNA sequencing allowed the sequencing and the comparison of the genomes from several individuals of the same species (Medini et al. 2005). This helped fuel the notion that an individual genome is insufficient to serve as an appropriate genomic reference, since it does not capture the diversity that represents the species. The idea emerged of a “pan-genome” that encompasses the genomic information of several representative individuals. Pan-genomics was initially applied to many smaller and simple genomes of microbial species, particularly to understand presence/absence variation (PAV) in genes (Medini et al. 2005). The idea of the pan-genome has since been applied to diverse species across all taxonomic kingdoms and has evolved to consider all possible variation present between genomes, including non-genic, PAV, copy number, and structural variation (Jayakodi et al. 2021). Pan-genomics has also been applied more broadly to groups of related species or genera, for “super pan-genomes.” While still in its infancy, pan-genomics of crop species can be particularly valuable for harnessing genomic variants and increasing rates of crop improvement. The application of pan-genomes in crop breeding is gaining increased interest due to the importance of food security and the need for more efficient and effective breeding methods. To date, pan-genomes have been applied to the improvement of various crops, including barley, maize, rice, tomato, and soybean (Gao et al. 2019; Gui et al. 2022; Jayakodi et al. 2020; Liu et al. 2020; Shang et al. 2022; Zhao et al. 2018). Applications of pan-genomics for wheat improvement have also become possible since the completion and the public release of multiple high-quality reference genomes (Walkowiak et al. 2020).
Wheat is a crucial crop globally, with widespread cultivation and significant economic importance, supplying a fifth of global calories and protein (Dixon 2007; Shiferaw et al. 2013). To maintain food security in the context of exponential growth of the human population while facing new challenges (e.g., global warming and climate change) in production, it is essential to create new wheat varieties with increased yield, better quality, and resistance or tolerance to abiotic and biotic stress (Abberton et al. 2016; Batley and Edwards 2016). Early wheat improvement relied on traditional breeding methods, where wheat lines were phenotypically selected in field trials, which is both costly and labor intensive. As our understanding of wheat genetics improved, it became possible to identify major effect genes underlying qualitative traits and to select for these genes through marker-assisted selection (MAS, see also Chap. 9). Marker-assisted selection has been successfully applied to certain traits, particularly disease resistance (Miedaner and Korzun 2012). Unfortunately, many key traits, including yield, have a complex and polygenic determinism. Selection of quantitative traits that are more complex and are influenced by non-genic features, several genes, or gene interactions, require more advanced tools for making DNA-based selections. With the recent availability of high-quality genome assembly and gene annotations for wheat, it has been possible to apply high-throughput genotyping arrays or genotype-by-sequencing methods to gather genome-wide variation information and select for these complex traits at the whole-genome level, through genomic selection (GS) (Haile et al. 2021). Nevertheless, identifying key major effect genes as well as the mechanisms underpinning more complex traits requires a deeper understanding of the diversity of wheat and the impact of genomic variation on phenotypic traits. It is critical to understand the diversity within wheat that is available to breeders in order to make breeding more efficient, identify suitable parents to use in targeted crosses, and select for the best possible combination of genes for rapid trait enhancement.
Despite its importance for food security, the application of genomics and pan-genomics for wheat improvement has been challenged by the large size and the complexity of its genome. The genome is composed of three separated diploid subgenomes, resulting in allohexaploidy (genome AABBDD), where the ‘A’ subgenome was derived from T. urartu, the ‘B’ subgenome from a species related to T. speltoides, and the ‘D’ genome from Ae. tauchii. The genome of modern bread wheat is estimated to be 17 gigabase-pairs (Gb) in length and is composed of ~ 90% repetitive elements. Recent achievements in genome sequencing and assembly technologies have enabled the release of multiple wheat genomes and tools to create a pan-genome, which is inspiring a new age of wheat breeding. In this review, we explore the concept of pan-genomes and a pan-genome of wheat, the history and evolution of the wheat genome and pan-genome, and the future outlook of wheat pan-genomics for research and applied breeding.
14.2 Motivations for Studying Pan-Genomes in Crop Breeding
During the last decade, there have been significant advancements in next-generation sequencing (NGS) technologies, which offer a direct view into DNA variation. These advancements have created numerous possibilities to investigate the connection between genotype and phenotype with greater precision than ever before. NGS has been used for various projects, including gene expression analysis, polymorphism detection, and the development of molecular markers (Barabaschi et al. 2012; Delseny et al. 2010). With the advent of affordable genome sequencing, breeders have started using NGS to sequence extensive groups of plants, which has enhanced the precision of identifying quantitative trait loci (QTL) and simplified the process of discovering genes. This has, in turn, formed the foundation for creating models to comprehend complex genotype–phenotype relationships at the whole-genome level. Over the past two decades, advancements in sequencing technologies, assembly techniques, and computational algorithms have enabled the release of genome sequences for over 700 plant species (Sun et al. 2022).
In parallel, advancements in using DNA-based tools for plant breeding, such as MAS and GS, have progressed significantly. Genomics approaches identified genomic markers associated with traits and were termed as QTL (Geldermann 1975). A single QTL can harbor many genes within the same locus (Beckmann and Soller 1983; Westman et al. 1997). MAS has been in use since the early 1990s and involves identifying genomic markers in silico, which are within causal genes for traits or are closely linked, which are then used to select individuals (Tanksley and Nelson 1996).
The development of reference genome assemblies has expedited the process of identifying candidate genes for in-demand traits. These assemblies serve as a basis for pinpointing single-nucleotide polymorphisms (SNPs), copy number variations (CNVs), and insertion–deletions (InDels) within an individual’s DNA sequence. The markers were used as the basis for conducting genome-wide association studies (GWAS) and genomic selection (GS), which involve comparing diversity panels with reference genomes to identify statistical associations between markers and traits (Crossa et al. 2017; Hayes and Goddard 2010; Varshney et al. 2009). Despite providing a greater insight into the diversity of plant species, particularly at the SNP level (Gore et al. 2009; McNally et al. 2009), reference genomes cover only a limited portion of the overall genomic space of a species and are inadequate in capturing variation across every individual within a given crop species (Bayer et al. 2020). A paradigm shift is occurring due to new advancements in genomics, which now take into account the significance and amount of structural variations present in the pan-genome of crop species. This includes capturing all types of SVs such as PAVs, CNVs, and repetitive elements or TEs, present throughout the entire genome of all individuals belonging to a plant species (Danilevicz et al. 2020; Golicz et al. 2016; Tao et al. 2019). By cataloging this variation and linking it to phenotypic/trait information, it is then possible select parents and candidate wheat lines in breeding programs with more advanced knowledge and decision support tools, allowing for more efficient and targeted crop improvement.
14.3 Historical Challenges and Progress in Wheat Genome Sequencing and Assembly
Prior to the availability of NGS, whole-genome sequencing was performed using the Sanger sequencing technology. Due to a combination of several factors, including the cost and low throughput of Sanger sequencing, and the size and complexity of some large genomes, many genomes were first cloned into bacterial artificial chromosomes (BACs) that included a few hundred thousand base-pairs per clone. This allowed for each BAC to be sequenced and assembled in parallel and then stitched together to assemble larger more complex genomes. After the release of the first human genome sequencing in 2000, which was achieved through the use of bacterial artificial chromosome (BAC) (Lander 2001; Venter et al. 2001), the Arabidopsis genome was the first plant genome to be sequenced using this approach. This was followed by the completion of multiple versions of the rice genome two years later (Goff et al. 2002; Yu et al. 2002). The wheat genome’s larger size, almost 40 times that of rice, and its complexity, which included a high proportion of repetitive sequences and homoeologous DNA copies from three subgenomes, made it economically unfeasible to employ a standard sequencing method. To tackle this challenge, the International Wheat Genome Sequencing Consortium (IWGSC) was established in 2005. The consortium divided the immense task among 20 countries based on chromosomes and chromosome arms. The approach employed genetic stocks that could be differentiated by flow cytometry on an individual chromosome basis (Consortium et al. 2014). Physical maps and minimum tiling paths were produced by fingerprinting BAC libraries, which were subsequently sequenced and assembled (Safár et al. 2010). Although the chromosome-by-chromosome approach was adopted, it took nearly ten years to implement and was only partially accomplished for few chromosomes, including chromosome 3B (Paux et al. 2008). Due to the large size of the hexaploid wheat genome, certain researchers have opted to pursue a different approach by focusing on the genomes of related diploid species, such as Ae. tauschii. This species has a much smaller genome size, approximately one-third of that of hexaploid wheat (~ 4.792 Gb) and does not have any interference from homoeologous DNA copies during physical mapping and eventual sequence assembly. Despite implementing this method, the initial use of regular agarose gels made the task seem overwhelming. However, to anchor contigs, higher throughput technologies such as SNaPshot BAC fingerprinting and Illumina Infinium SNP array were utilized. It took a decade to produce the first version of the Ae. tauschii physical map, which involved fingerprinting 461,706 BAC clones and assembling them into 2263 contigs. Afterward, 7185 molecular markers were utilized to anchor these contigs onto a genetic map (Luo et al. 2013). Despite some success with Ae. tauschii, the BAC approach had limited achievement in hexaploid wheat and the approach was slowly abandoned for wheat once more advanced DNA sequencing, sequencing library preparation, and genome assembly technologies became available.
In the 2000s, wheat genome sequencing was boosted by Illumina sequencing technologies, which were able to perform short read paired-end sequencing at high depth and low cost. The sequencing was first done on the diploid ancestors of common wheat due to their smaller genome size and early challenges of applying short read data to large polyploidy genomes. The draft genome assembly for Ae. tauschii, the D genome donor for bread wheat, was completed using short read sequencing methods to about 90 × coverage (Jia et al. 2013). Approximately, 83.4% of the genome was covered by the assembled scaffolds, and out of these, 65.9% were identified as transposable elements (TEs). Using RNA-seq data from different tissues, a total of 43,150 protein-encoding genes were identified. A comparable approach was employed to construct the genome sequence of the A genome contributor, T. urartu. The assembly that was obtained had a total length of 3.92 Gb, which corresponds to 79.35% of the estimated size of the A genome (4.94 Gb). However, due to subgenome interactions and evolutionary processes spanning around 10,000 years, the genomes of the progenitors are not able to fully depict their counterparts in the common wheat genome. Therefore, the sequencing of the common wheat genome was yet to be achieved.
The first sequencing of the common wheat genome for the landrace CHINESE SPRING was accomplished using Roche 454 pyrosequencing, specifically the GS FLX Titanium and GS FLX1 platforms, which were used to sequence the wheat genome to about 5 × coverage. Sequencing of related progenitors was also performed using various platforms, such as Illumina methods for sequencing of T. monococcum, the A genome donor of bread wheat. Likewise, Ae. tauschii was sequenced using the Roche 454 sequencing platform. While whole-genome data was not yet available, cDNA sequences were sequenced from Ae. speltoides, which has a genome similar to the B genome. Using the SOLiD sequencing platform, additional short reads of CHINESE SPRING were generated. These yielded 95,000 predicted gene models, with most of them designated to either the A, B, or D subgenome. Despite its high degree of fragmentation, the draft genome was still considered valuable, as it was the first wheat genome available for community use (Brenchley et al. 2012).
As the IWGSC adopted the chromosome-based BAC sequencing approach, progress was consistently made. As NGS became available, it was possible to sequence the BACs using more high-throughput methods. The approach involved developing sequencing libraries from the DNA of individual chromosomes or their arms and subsequently sequencing pair-end reads on the Illumina HiSeq 2000 platform. The assembly obtained, which resembled the 454 assembly, comprised approximately 500,000 contigs with N50 values ranging from 1.7 to 8.9 kb. Its total size was 10.2 Gb. These contigs, taken together, make up 61% of the estimated hexaploid wheat genome. Predictions were made for a total of 133,090 high confidence genes, as well as 890,576 low confidence genes. Using a genetic map, just over half of the high confidence genes were assigned genetic positions (Mascher et al. 2013), allowing them to be considered within the context of the telosome-based assembly resources for each chromosome arm. This led to the completion of a draft genome assembly of wheat, known as the IWGSC chromosome survey sequence (CSS) assembly (Consortium et al. 2014).
The IWGSC also accomplished a noteworthy feat when they generated a reference-level sequence of chromosome 3B (Choulet et al. 2014). This high-quality sequence was created using a minimum tiling path consisting of 8452 BACs, spanning 774 Mb, and containing 5326 protein-coding genes as well as 85% of TEs. Additionally, a molecular-genetic map (CHINESE SPRING x RENAN) was used for long-range orientation of DNA sequences. The assembly of chromosome 3B demonstrated the success of the chromosome-based BAC sequencing strategy, although the assembly remained approximately 7% incomplete.
14.4 The Completion of a Chromosome-Scale Assembly of Hexaploid Wheat
While evidence suggested the BAC sequencing approach could work for achieving a chromosome-based wheat genome assembly, the complexity of the genome, high repeat content, high transposon activity, large genome size, and allopolyploidy were continuing to hamper assembly efforts. Meanwhile, third-generation sequencing technologies, which were created by Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT), surfaced and progressed quickly. These techniques produce reads with substantially longer lengths and have been extensively employed, in combination with established assembly algorithms, to construct intricate and sizable plant genomes with unparalleled precision (Cheng et al. 2021; Koren et al. 2017; Niu et al. 2022). This led to a paradigm shift away from BAC sequencing and toward the direct shotgun sequencing of the genome using more advanced sequencing technologies and assembly algorithms.
A new assembly method called MaSuRCA was used to assemble wheat using a hybrid approach that combined the strengths of both PacBio long reads, which have high error rates, and Illumina short reads, which are more accurate. This method was initially used to create a genome assembly of Ae. tauschii (Zimin et al. 2017a). To obtain a comprehensive sequence coverage of the genome, a combination of sequencing methods was employed, including over 19 million PacBio reads providing approximately 38 × coverage of the D genome, 177 × coverage from Illumina HiSeq 2500 reads consisting of 200-base paired-end reads, and MiSeq reads consisting of 250-base paired-end reads. The sequencing libraries with a range of insert sizes yielded a total coverage of 200 × of the genome. The genome’s quality was validated through a comparison with optical maps and BAC assemblies that were produced independently. Subsequently, the pipeline was utilized to produce the initial near-complete hexaploid wheat genome for CHINESE SPRING (Zimin et al. 2017b). Triticum 1.0 was a genome assembly consisting of 829,839 contigs with a total size of 17.05 Gb, with a contig and scaffold N50 of 76.3 kb and 101.2 kb, respectively. Another method involved assembling long reads directly with the FALCON assembler, which produced FALCON Trit1.0 with a size of 12.94 Gb. Although this version was shorter than the MaSuRCA-assembled version, it had a longer contig N50 of 215.3 kb. Using the genome alignment tool MUMmer (Kurtz et al. 2004), the combination of Triticum 1.0 and Trit1.0 resulted in a final assembly that spans almost the entire wheat genome, with a size of 15.3 Gb and a contig N50 of 232.6 kb.
At the same time, an alternative approach was also taken to create the CHINESE SPRING genome assembly using short reads (Clavijo et al. 2017). The approach involved 1.1 billion 250-bp paired-end reads (33 × genome coverage) from CHINESE SPRING short insert libraries, and 68 × coverage of long insert libraries, yielding the TGACv1 version of the wheat genome assembly. This version spanned 13.43 Gb and accounted for over 78% of the wheat genome. In addition to the improved assembly, strand-specific Illumina RNA-seq and PacBio full-length cDNAs were combined to achieve better annotation. Although chromosome-level assembly was not attained, this new wheat genome assembly was now available for the broader scientific community to utilize, bringing the prospect of a high-quality reference genome into focus.
Shortly thereafter, a breakthrough was made with the release of new short read assemblers. NRGene’s DeNovoMagic (NRGene, Ness Ziona, Israel) algorithm and the TRITEX pipeline (Monat et al. 2019) for short read assemblies demonstrated that a shotgun whole-genome sequencing approach could be achieved when combining different Illumina library sizes and preparation methods. The AABB genome of wild emmer wheat (WEW), which represents the reference-level genome of polyploid wheat, was produced through the utilization of the DeNovoMagic algorithm (Avni et al. 2017). By sequencing on Illumina HiSeq 2500 machines, a total of 2.1 Terabase-pairs were generated, comprising 176 × genome coverage reads from five libraries. The insert sizes in the libraries ranged from 450 bp to 10 kb. The scaffolds were then consolidated using a high-density molecular-genetic linkage map and additional reads from a three-dimensional (3D) conformation capture Hi-C library. Ultimately, the final assembly was 10.5 Gb, accounting for 87.5% of the predicted tetraploid wheat genome. The annotation of 110,544 gene models provided strong evidence for the high quality of this genome assembly. Among these models, 58.8% (65,012) were identified as high confidence gene models, while the remaining 41.2% were of low confidence. This assembly successfully captured 98.4% of the total expected gene sets of WEW, as verified by BUSCO (Simão et al. 2015). Additionally, it was utilized for identifying the genes that played a role in the early domestication of wheat, as reported by Avni et al. (2017). After the completion of the WEW genome, bread wheat genome sequencing efforts quickly pivoted toward the same shotgun genomics approach. The successful completion of the bread wheat genome IWGSC RefSeq v1.0 was achieved using a combination of similar techniques and software. According to Consortium et al. (2018), DeNovoMAGIC2 utilized the complete genome as the primary framework and incorporated various sources of data such as physical maps, genotyping-by-sequencing data, and Hi-C data. The common wheat genome was assembled into 21 pseudomolecules at the chromosome scale, which were assigned to the subgenomes A, B, and D. This resulted in a genome assembly with a super-scaffold N50 of 22.8 Mb, and total length of 14.5 Gb. Using a similar assembly approach, the genome sequencing of durum wheat (DW) was completed shortly after (Maccaferri et al. 2019).
14.5 Progress Toward a Wheat Pan-Genome
In 2018, the IWGSC released the first reference-quality genome sequence for the wheat landrace CHINESE SPRING, which marked a significant change in the use of genomics as a research tool for wheat. The publication enabled the wider research community to have easy access to this tool (Consortium et al. 2018). The CHINESE SPRING genome assembly was a major milestone in wheat genomics research, and within a few years, it has already laid the foundation for countless studies dissecting the genome to understand wheat biology. However, CHINESE SPRING shares only a distant ancestral connection with the majority of current wheat varieties. Additionally, due to the considerable diversity present within the species, a single genome sequence is insufficient for fully representing its genetic makeup. Additional pan-genome information is required to identify new genetic diversity that can enhance traits and understand the mechanism behind the traits present in elite wheat cultivars. Fortunately, with new short-read assembly algorithms capable of shotgun sequencing, the path forward to additional genomes would no longer be a technical limitation.
Choosing crop genotypes for pan-genome analysis is a challenging task as the objective is to encompass a wide range of genetic variations using a limited number of representative genotypes for the particular species. This selection procedure necessitates the acquisition of genome-wide genotypic data from either entire genebank collections or representative subgroups that cover all significant germplasm groups within the species. Recent reports have described several genebank genomics studies on rice (Wang et al. 2018), barley (Milner et al. 2019), and wheat (Juliana et al. 2019). Soleimani et al. (2020) have described different methods that can be used to choose core sets for pan-genome analysis. One tool that aims to maximize diversity, representativeness, and allelic richness of core sets is Core Hunter (De Beukelaer et al. 2018). It achieves this by using various algorithms that operate on genetic distance matrices. To further customize the selection process, clustering of the diversity space through principal component analysis (Patterson et al. 2006) or model-based ancestry estimation (Alexander et al. 2009) can be used. Pan-genome panels offer the possibility of incorporating not only cultivated plant varieties but also wild progenitors or ancestors of polyploid species. For example, teosinte as a wild progenitor of maize and wild emmer or Aegilops tauschii as progenitors of wheat. These wild relatives are valuable out-groups and represent diversity available in the secondary and tertiary gene pools. These relatives could be used to determine the ancestral states for SVs or because of their significance in introgression breeding (Harlan and de Wet 1971). Besides emphasizing on incorporating diverse global varieties in a crop, a pan-genome initiative might also choose specific genotypes that have a significant role in breeding and genetics. These could comprise founder genotypes of breeding programs, experimental population parents (Yu et al. 2008), or genotypes that can be genetically modified (Jain et al. 2019; Schreiber et al. 2020) to optimize the advantages for both research and breeding communities. These chosen accessions will serve as reference genotypes for future functional and genetic studies in pan-genomic research.
The International 10 + Wheat Genomes Project (www.10wheatgenomes.com) was established in 2019 with the goal of creating reference-quality genome assemblies for at least ten diverse bread wheat cultivars. Using genomic diversity analysis of 3800 wheat samples, ten wheat lines were chosen and sequenced utilizing Illumina short read sequencing technologies, and then assembled using NRGene’s DeNovoMagic algorithm (NRGene, Ness Ziona, Israel). Subsequently, all these assemblies were organized into subgenome-aware pseudomolecules with the aid of Hi-C technology (van Berkum et al. 2010). Additionally, five other wheat varieties were also sequenced and assembled to the scaffold level using separate short-read assembly algorithms established at the Earlham Institute (Norwich, UK).
A gene projection strategy was implemented and applied to all assemblies to evaluate and compare the gene content of the newly sequenced lines in a fair and consistent manner, given the lack of genome-specific transcriptome data available at that time. This strategy involved using the CHINESE SPRING reference gene models and transferring them to all assemblies. Differences in gene content among the 10 + wheat reference genomes were observed, likely due to the complex breeding histories of the selected lines. These variations in gene content were found to be linked with adaptation to different environments and with efforts to enhance grain yield, quality, and resistance to abiotic and biotic stresses. Significant structural rearrangements and introgressions from wild relatives were observed upon comparing the pseudomolecule structures of the reference sequences. This underscores the importance of having multiple reference genomes of quality (at pseudomolecule level) instead of relying on resequencing approaches, as only chromosome-level assemblies can provide information on large- and small-scale structural rearrangements with a high degree of resolution and accuracy. The study conducted by Walkowiak et al. (2020) illustrates how the wheat pan-genomes can be utilized to study causal genes for traits, as the genomes were used to uncover the gene Sm1, known for conferring resistance against midge. With the availability of recently sequenced and compiled wheat reference genomes, there is an unprecedented opportunity to identify functional genes and enhance wheat breeding. The subsequent phase of the project will involve generating de novo gene predictions for all chromosome-scale assemblies using extensive transcriptome data. These data will offer a comprehensive understanding of the functional and regulatory arrangement of the wheat pan-genome.
While the 10 + Wheat Genomes Project provided the first insights into the wheat pan-genome, sequencing and assembly methods continued to evolve. Throughput increased for both PacBio and ONT sequencing platforms, leading to additional genome assemblies (Aury et al. 2022). Further, PacBio released its HiFi sequencing method based on circular consensus sequencing, which significantly improved sequencing and assembly accuracy. These long and accurate sequencing reads have led to the highest-quality genome assemblies of wheat achieved thus far. With the upcoming release of new long read sequencing technologies with high accuracy and output, such as the Revio platform from PacBio, it is expected that additional genomes for wheat will be released in the coming years. While no longer constrained by technological limitations in genome sequencing and assembly, the next chapter begins for integrating these data into a functional pan-genome that will drive future research and breeding.
14.6 A Functional Pan-Genome for Wheat Research and Applied Breeding
Pan-genome construction is the process of creating a comprehensive set of genetic information from a collection of related genomes. It is a complex task, requiring the use of multiple approaches and techniques. It involves assembling and annotating all genomic information and variants, can be used to understand genome and gene evolution, discover new genes and alleles, and investigate gene–gene interaction networks.
To construct a pan-genome, two primary methods can be utilized, whole-genome assembly and comparative genomics. Whole-genome assembly involves assembling all of the reads from a collection of genomes into a single, contiguous genome. The steps for whole-genome assembly are well-documented (Jung et al. 2020). The approach is most appropriate for genomes that are closely related and possess significant sequence similarity. It offers the benefit of an all-encompassing perspective on the species’ genetic variation, but it is often restricted by the number of genomes that can be sequenced. Comparative genomics (Pop et al. 2004), on the other hand, involves comparing and contrasting multiple genomes to identify shared and unique components. This method is most suitable for more distantly related genomes with lower sequence similarity.
The ability to assemble high-quality reference genomes for numerous plants simultaneously has been made possible by recent advancements in sequencing technologies and bioinformatic tools. Despite this progress, it is still challenging to perform combined analysis of multiple genomes or a subset of genomes and provide readily accessible genetic information to end-users, such as researchers and breeders (Li et al. 2020b). The comparison, analysis, and visualization of multiple reference genomes and their diversity necessitate powerful and specialized computational strategies and tools. De novo assembly, iterative assembly, and graph-based assembly methods have been employed to construct pan-genomes (Li et al. 2014; Liu and Tian 2020).
14.6.1 De Novo Assembly
Constructing a pan-genome can be achieved through the de novo assembly of genomes from multiple individuals, followed by comparative analysis to identify variant types and classify them as core or flexible genome components. This approach has been discussed by Mahmoud et al. (2019). Technological advancements in sequencing and assembly methods have enabled the generation of high-quality, chromosome-level plant genomes, including telomere-to-telomere genome assemblies (Miga et al. 2020). However, generating accurate genome assemblies can be costly, especially for large plant genomes, and may not be practical when dealing with hundreds of reference genomes for a single species (Hurgobin and Edwards 2017). Nevertheless, the 10 + Wheat Genomes Project was successful at the construction of several chromosome-scale assemblies. Along with these genomes were tools to visualize haplotype blocks representing shared or unique regions between the assemblies (http://www.crop-haplotypes.com/) (Brinton et al. 2020). Likewise, many of the wheat genomes had major introgressions or large structural variants, which could be visualized using synteny viewers (https://kiranbandi.github.io/10wheatgenomes/, http://10wheatgenomes.plantinformatics.io/).
14.6.2 Iterative Assembly
The iterative assembly approach differs from de novo assembly in that it commences with the creation of a single-reference genome, which is then used as a framework for the sequential alignment of reads from other samples. Any unmapped reads are subsequently assembled and incorporated into the reference genome to form a non-redundant pan-genome (Golicz et al. 2016). This technique is less expensive than de novo assembly since low sequencing depths can be used for each sample, allowing for the pooling of numerous samples. Nevertheless, the iterative assembly method may struggle to handle genomes that contain many repeat regions and is not capable of detecting large structural variations that cannot be covered by individual short reads (Jiao and Schneeberger 2017). Resequencing and iterative assembly methods have been applied to wheat (Montenegro et al. 2017; Watson-Haigh et al. 2018). However, evidence suggests that wheat has a very plastic genome due to its allopolyploidy and has abundant PAV, CNV, and SV that are important for trait variation www.10wheatgenomes.com, (Nilsen et al. 2020). Therefore, iterative assembly approaches, particularly low-coverage reference-based analyses, are highly limiting when exploring wheat pan-genomics.
14.6.3 Graph-Based Assembly
Pan-genomes can also be constructed using graphs. The most commonly used graph for this purpose is the compacted de Bruijn graph, which integrates genetic information from different accessions of a species (Chikhi et al. 2016; Li et al. 2020a). In contrast, the bi-directed variation graphs capture genetic variations throughout a population and identify their potential positions on a reference genome. Compared to traditional linear genomes, graph-based pan-genomes have been shown to significantly mitigate reference bias (Garrison et al. 2018). However, graph-based pan-genomes are challenging to construct and apply due to several factors, including the intricate nature of plant genomes with their high repeat content and polyploidy. Additionally, there is a shortage of common downstream analysis tools and visualization techniques for the graph, which further adds to the limitations. Despite these challenges, graph-based genomes have strengths compared to other methods and may have more widespread applications for wheat research and breeding in the future, particularly as tools for graph-based assembly of more complex genomes improve.
14.6.4 Pan-Genome Annotation and Other Pan-Omics
Once the pan-genome has been assembled, there are several techniques that can be used to annotate it. One technique is to use gene prediction software to identify genes in the pan-genome. This can be done using homology-based or de novo gene prediction algorithms. There is a plethora of ab initio gene prediction software (Scalzitti et al. 2020), including Augustus (Stanke and Morgenstern 2005), Genscan (Burge and Karlin 1997), GeneID (Parra et al. 2000), GlimmerHMM (Majoros et al. 2004), and Snap (Korf 2004). Another technique to annotate the pan-genome is to use comparative genomics to identify conserved or novel gene families. This involves comparing the genomes of different species to identify shared and unique components. By comparing gene sequences between two species, it is possible to identify regions of similarity that may indicate similar functions. In wheat, comparative genomics has been used for identifying resistance genes (Marchal et al. 2020) and uncovering the molecular basis of nitrogen-use efficiency (Shi et al. 2022). In addition to annotating the gene space, there is increasing interest in expanding the annotation of the pan-genome to include the dynamics of gene expression (pan-transcriptomics), epigenomic modifications (epipan-genomics), as well as interaction networks between variants as well as genes, and associating these directly with biological traits. Such a complete atlas of biological information will equip researchers and breeders with unprecedented tools for wheat research and improvement.
14.6.5 Applying the Pan-Genome to Breeding
After constructing and annotating the pan-genome, the subsequent step involves utilizing it for crop enhancement. The effectiveness of next-generation breeding technologies, such as transgenics and CRISPR-Cas9 gene editing, has been proven for wheat (Nilsen et al. 2020). However, regulatory challenges exist that may limit the widespread adoption of these methods for delivering new wheat cultivars. As a result, wheat breeding will likely involve generating biparental populations and screening for progeny for some time to come. Gene discovery has certainly benefitted from the availability of pan-genomics resources for wheat, facilitating marker discovery that can be applied to MAS and making screening of parental lines and progeny more efficient (www.10wheatgenomes.com). With the availability of more genome assemblies that are representative of the genes and genomic variants that can be used in breeding, the need to generate additional high-quality genomes will likely lessen as genomes can be imputed based on lower coverage haplotype information; for example, from genotype-by-sequencing or high-throughput SNP arrays (Alipour et al. 2019). Having genomic information available for the parental materials being used in crosses, even if imputed, will allow for breeders to make stronger associations between traits of interests and variants within the genome, allowing for more efficient and targeted genomic-based selections to be made in their resulting progeny through GS.
14.7 Conclusion and Future Directions
Owing to its ability to identify novel genetic variations that can enhance crucial traits, the pan-genome serves as a valuable asset for crop breeding, specifically in wheat. Through consistent pan-genome research in crops, more robust and productive varieties are expected to be developed, resulting in benefits for farmers and consumers worldwide. While it is difficult to predict all possible future applications of pan-genomics to wheat breeding, the resources are now available to innovate. With recent advances in GS, artificial intelligence, and deep learning, one can only imagine the possibilities when applying these tools to pan-genomics, particularly if the pan-genomes are well annotated and have associated phenotypic data generated through applied breeding. This may not only be able to predict the performance of parents or offspring but could potentially help optimize designer genomes for specific purposes, environments, or stresses.
References
Abberton M, Batley J, Bentley A, Bryant J, Cai H, Cockram J, Costa de Oliveira A, Cseke LJ, Dempewolf H, De Pace C, Edwards D, Gepts P, Greenland A, Hall AE, Henry R, Hori K, Howe GT, Hughes S, Humphreys M, Lightfoot D, Marshall A, Mayes S, Nguyen HT, Ogbonnaya FC, Ortiz R, Paterson AH, Tuberosa R, Valliyodan B, Varshney RK, Yano M (2016) Global agricultural intensification during climate change: a role for genomics. Plant Biotechnol J 14:1095–1098
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Alipour H, Bai G, Zhang G, Bihamta MR, Mohammadi V, Peyghambari SA (2019) Imputation accuracy of wheat genotyping-by-sequencing (GBS) data using barley and wheat genome references. PLoS ONE 14:e0208614
Aury J-M, Engelen S, Istace B, Monat C, Lasserre-Zuber P, Belser C, Cruaud C, Rimbert H, Leroy P, Arribat S, Dufau I, Bellec A, Grimbichler D, Papon N, Paux E, Ranoux M, Alberti A, Wincker P, Choulet F (2022) Long-read and chromosome-scale assembly of the hexaploid wheat genome achieves high resolution for research and breeding. GigaScience 11
Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, Hale I, Mascher M, Spannagl M, Wiebe K, Jordan KW, Golan G, Deek J, Ben-Zvi B, Ben-Zvi G, Himmelbach A, MacLachlan RP, Sharpe AG, Fritz A, Ben-David R, Budak H, Fahima T, Korol A, Faris JD, Hernandez A, Mikel MA, Levy AA, Steffenson B, Maccaferri M, Tuberosa R, Cattivelli L, Faccioli P, Ceriotti A, Kashkush K, Pourkheirandish M, Komatsuda T, Eilam T, Sela H, Sharon A, Ohad N, Chamovitz DA, Mayer KFX, Stein N, Ronen G, Peleg Z, Pozniak CJ, Akhunov ED, Distelfeld A (2017) Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science (New York, NY) 357:93
Barabaschi D, Guerra D, Lacrima K, Laino P, Michelotti V, Urso S, Valè G, Cattivelli L (2012) Emerging knowledge from genome sequencing of crop species. Mol Biotechnol 50:250–266
Batley J, Edwards D (2016) The application of genomics and bioinformatics to accelerate crop improvement in a changing climate. Curr Opin Plant Biol 30:78–81
Bayer PE, Golicz AA, Scheben A, Batley J, Edwards D (2020) Plant pan-genomes are the new reference. Nature Plants 6:914–920
Beckmann JS, Soller M (1983) Restriction fragment length polymorphisms in genetic improvement: methodologies, mapping and costs. Theor Appl Genet 67:35–43
Brenchley R, Spannagl M, Pfeifer M, Barker GLA, D’Amore R, Allen AM, McKenzie N, Kramer M, Kerhornou A, Bolser D, Kay S, Waite D, Trick M, Bancroft I, Gu Y, Huo N, Luo M-C, Sehgal S, Gill B, Kianian S, Anderson O, Kersey P, Dvorak J, McCombie WR, Hall A, Mayer KFX, Edwards KJ, Bevan MW, Hall N (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491:705
Brinton J, Ramirez-Gonzalez RH, Simmonds J, Wingen L, Orford S, Griffiths S, Haberer G, Spannagl M, Walkowiak S, Pozniak C, Uauy C, Wheat Genome P (2020) A haplotype-led approach to increase the precision of wheat breeding. Communications Biology 3:712
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. Journal of molecular biology 268
Cheng H, Concepcion GT, Feng X, Zhang H, Li H (2021) Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods 18:170–175
Chikhi R, Limasset A, Medvedev P (2016) Compacting de Bruijn graphs from sequencing data quickly and in low memory. Bioinformatics 32:i201–i208
Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, Pingault L, Sourdille P, Couloux A, Paux E, Leroy P, Mangenot S, Guilhot N, Le Gouis J, Balfourier F, Alaux M, Jamilloux V, Poulain J, Durand C, Bellec A, Gaspin C, Safar J, Dolezel J, Rogers J, Vandepoele K, Aury J-M, Mayer K, Berges H, Quesneville H, Wincker P, Feuillet C (2014) Structural and functional partitioning of bread wheat chromosome 3B. Science (New York, NY) 345:1249721
Clavijo BJ, Venturini L, Schudoma C, Accinelli GG, Kaithakottil G, Wright J, Borrill P, Kettleborough G, Heavens D, Chapman H, Lipscombe J, Barker T, Lu F-H, McKenzie N, Raats D, Ramirez-Gonzalez RH, Coince A, Peel N, Percival-Alwyn L, Duncan O, Trösch J, Yu G, Bolser DM, Namaati G, Kerhornou A, Spannagl M, Gundlach H, Haberer G, Davey RP, Fosker C, Palma FD, Phillips AL, Millar AH, Kersey PJ, Uauy C, Krasileva KV, Swarbreck D, Bevan MW, Clark MD (2017) An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res 27:885–896
Consortium IWGS, Mayer KF, Rogers J, Doležel J, Pozniak C, Eversole K, Feuillet C, Gill B, Friebe B, Lukaszewski AJ (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science (New York, NY) 345:1251788
Consortium IWGS, Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science (New York, NY) 361:eaar7191
Crossa J, Pérez-Rodríguez P, Cuevas J, Montesinos-López O, Jarquín D, De Los CG, Burgueño J, González-Camacho JM, Pérez-Elizalde S, Beyene Y (2017) Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci 22:961–975
Danilevicz MF, Tay Fernandez CG, Marsh JI, Bayer PE, Edwards D (2020) Plant pangenomics: approaches, applications and advancements. Curr Opin Plant Biol 54:18–25
De Beukelaer H, Davenport GF, Fack V (2018) Core Hunter 3: flexible core subset selection. BMC Bioinformatics 19:203
Delseny M, Han B, Hsing Y (2010) High throughput DNA sequencing: the new sequencing revolution. Plant Science: An International Journal of Experimental Plant Biology 179:407–422
Dixon J (2007) The economics of wheat; Research challenges from field to fork. Wheat production in stressed environments. In: Proceedings of international wheat conference, 7; Mar de Plata (Argentina); 27 Nov–2 Dec 2005. ^ TWheat production in stressed environments Proceedings of International Wheat Conference, 7; Mar de Plata (Argentina); 27 Nov-2 Dec 2005^ ABuck, HT Nisi, JE Salomon, N^ ADordrecht (Netherlands)^ BSpringer^ C2007
Gao L, Gonda I, Sun H, Ma Q, Bao K, Tieman DM, Burzynski-Chang EA, Fish TL, Stromberg KA, Sacks GL, Thannhauser TW, Foolad MR, Diez MJ, Blanca J, Canizares J, Xu Y, van der Knaap E, Huang S, Klee HJ, Giovannoni JJ, Fei Z (2019) The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat Genet 51:1044–1051
Garrison E, Sirén J, Novak AM, Hickey G, Eizenga JM, Dawson ET, Jones W, Garg S, Markello C, Lin MF, Paten B, Durbin R (2018) Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat Biotechnol 36:875–879
Geldermann H (1975) Investigations on inheritance of quantitative characters in animals by gene markers I. Methods. Theoretical and Applied Genetics 46:319–330
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun WL, Chen L, Cooper B, Park S, Wood TC, Mao L, Quail P, Wing R, Dean R, Yu Y, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller RM, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science (New York, NY) 296:92–100
Golicz AA, Bayer PE, Barker GC, Edger PP, Kim H, Martinez PA, Chan CKK, Severn-Ellis A, McCombie WR, Parkin IA (2016) The pangenome of an agronomically important crop plant Brassica oleracea. Nat Commun 7:1–8
Gore MA, Chia J-M, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, Ware DH, Buckler ES (2009) A first-generation haplotype map of maize. Science (New York, NY) 326:1115–1117
Gui S, Wei W, Jiang C, Luo J, Chen L, Wu S, Li W, Wang Y, Li S, Yang N, Li Q, Fernie AR, Yan J (2022) A pan-Zea genome map for enhancing maize improvement. Genome Biol 23:178
Haile TA, Walkowiak S, N’Diaye A, Clarke JM, Hucl PJ, Cuthbert RD, Knox RE, Pozniak CJ (2021) Genomic prediction of agronomic traits in wheat using different models and cross-validation designs. Theor Appl Genet 134:381–398
Harlan JR, de Wet JMJ (1971) Toward a rational classification of cultivated plants. Taxon 20:509–517
Hayes B, Goddard M (2010) Genome-wide association and genomic selection in animal breeding. Genome 53:876–883
Hurgobin B, Edwards D (2017) SNP discovery using a pangenome: has the single reference approach become obsolete? Biology 6(1):21. https://doi.org/10.3390/biology6010021. PMID: 28287462; PMCID: PMC5372014
Jain R, Jenkins J, Shu S, Chern M, Martin JA, Copetti D, Duong PQ, Pham NT, Kudrna DA, Talag J, Schackwitz WS, Lipzen AM, Dilworth D, Bauer D, Grimwood J, Nelson CR, Xing F, Xie W, Barry KW, Wing RA, Schmutz J, Li G, Ronald PC (2019) Genome sequence of the model rice variety KitaakeX. BMC Genomics 20:905
Jayakodi M, Padmarasu S, Haberer G, Bonthala VS, Gundlach H, Monat C, Lux T, Kamal N, Lang D, Himmelbach A, Ens J, Zhang X-Q, Angessa TT, Zhou G, Tan C, Hill C, Wang P, Schreiber M, Boston LB, Plott C, Jenkins J, Guo Y, Fiebig A, Budak H, Xu D, Zhang J, Wang C, Grimwood J, Schmutz J, Guo G, Zhang G, Mochida K, Hirayama T, Sato K, Chalmers KJ, Langridge P, Waugh R, Pozniak CJ, Scholz U, Mayer KFX, Spannagl M, Li C, Mascher M, Stein N (2020) The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 588:284–289
Jayakodi M, Schreiber M, Stein N, Mascher M (2021) Building pan-genome infrastructures for crop plants and their use in association genetics. DNA Research 28:dsaa030
Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, Jing R, Zhang C, Ma Y, Gao L, Gao C, Spannagl M, Mayer KFX, Li D, Pan S, Zheng F, Hu Q, Xia X, Li J, Liang Q, Chen J, Wicker T, Gou C, Kuang H, He G, Luo Y, Keller B, Xia Q, Lu P, Wang J, Zou H, Zhang R, Xu J, Gao J, Middleton C, Quan Z, Liu G, Wang J, International Wheat Genome Sequencing C, Yang H, Liu X, He Z, Mao L, Wang J (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496:91
Jiao W-B, Schneeberger K (2017) The impact of third generation genomic technologies on plant genome assembly. Curr Opin Plant Biol 36:64–70
Juliana P, Poland J, Huerta-Espino J, Shrestha S, Crossa J, Crespo-Herrera L, Toledo FH, Govindan V, Mondal S, Kumar U, Bhavani S, Singh PK, Randhawa MS, He X, Guzman C, Dreisigacker S, Rouse MN, Jin Y, Pérez-Rodríguez P, Montesinos-López OA, Singh D, Mokhlesur Rahman M, Marza F, Singh RP (2019) Improving grain yield, stress resilience and quality of bread wheat using large-scale genomics. Nature Genetics
Jung H, Ventura T, Chung JS, Kim W-J, Nam B-H, Kong HJ, Kim Y-O, Jeon M-S, Eyun S-i (2020) Twelve quick steps for genome assembly and annotation in the classroom. PLoS Comput Biol 16:e1008325
Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736
Korf I (2004) Gene finding in novel genomes. BMC Bioinformatics 5:59
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5:1–9
Lander ES (2001) Initial sequencing and analysis of the human germane. Nature 409:860–921
Li H, Feng X, Chu C (2020a) The design and construction of reference pangenome graphs with minigraph. Genome Biol 21:1–19
Li H, Feng X, Chu C (2020b) The design and construction of reference pangenome graphs with minigraph. Genome Biol 21:265
Li Y-h, Zhou G, Ma J, Jiang W, Jin L-g, Zhang Z, Guo Y, Zhang J, Sui Y, Zheng L, Zhang S-s, Zuo Q, Shi X-h, Li Y-f, Zhang W-k, Hu Y, Kong G, Hong H-l, Tan B, Song J, Liu Z-x, Wang Y, Ruan H, Yeung CKL, Liu J, Wang H, Zhang L-j, Guan R-x, Wang K-j, Li W-b, Chen S-y, Chang R-z, Jiang Z, Jackson SA, Li R, Qiu L-j (2014) De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat Biotechnol 32:1045–1052
Liu Y, Du H, Li P, Shen Y, Peng H, Liu S, Zhou GA, Zhang H, Liu Z, Shi M, Huang X, Li Y, Zhang M, Wang Z, Zhu B, Han B, Liang C, Tian Z (2020) Pan-genome of wild and cultivated soybeans. Cell 182:162-176.e113
Liu Y, Tian Z (2020) From one linear genome to a graph-based pan-genome: a new era for genomics. Science China Life Sciences 63:1938–1941
Luo MC, Gu YQ, You FM, Deal KR, Ma Y, Hu Y, Huo N, Wang Y, Wang J, Chen S, Jorgensen CM, Zhang Y, McGuire PE, Pasternak S, Stein JC, Ware D, Kramer M, McCombie WR, Kianian SF, Martis MM, Mayer KF, Sehgal SK, Li W, Gill BS, Bevan MW, Simkova H, Dolezel J, Weining S, Lazo GR, Anderson OD, Dvorak J (2013) A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci USA 110:7940–7945
Maccaferri M, Harris NS, Twardziok SO, Pasam RK, Gundlach H, Spannagl M, Ormanbekova D, Lux T, Prade VM, Milner SG (2019) Durum wheat genome highlights past domestication signatures and future improvement targets. Nat Genet 51:885–895
Mahmoud M, Gobet N, Cruz-Dávalos DI, Mounier N, Dessimoz C, Sedlazeck FJ (2019) Structural variant calling: the long and the short of it. Genome Biol 20:1–14
Majoros WH, Pertea M, Salzberg SL (2004) TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20:2878–2879
Marchal C, Wheat Genome P, Haberer G, Spannagl M, Uauy C (2020) Comparative genomics and functional studies of wheat BED-NLR loci. Genes (Basel) 11
Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, Muñoz-Amatriaín M, Close TJ, Wise RP, Schulman AH, Himmelbach A, Mayer KFX, Scholz U, Poland JA, Stein N, Waugh R (2013) Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J 76:718–727
McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, Stokowski R, Ballinger DG, Frazer KA, Cox DR, Padhukasahasram B, Bustamante CD, Weigel D, Mackill DJ, Bruskiewich RM, Rätsch G, Buell CR, Leung H, Leach JE (2009) Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci 106:12273–12278
Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R (2005) The microbial pan-genome. Curr Opin Genet Dev 15:589–594
Miedaner T, Korzun V (2012) Marker-assisted selection for disease resistance in wheat and barley breeding. Phytopathology® 102:560–566
Miga KH, Koren S, Rhie A, Vollger MR, Gershman A, Bzikadze A, Brooks S, Howe E, Porubsky D, Logsdon GA, Schneider VA, Potapova T, Wood J, Chow W, Armstrong J, Fredrickson J, Pak E, Tigyi K, Kremitzki M, Markovic C, Maduro V, Dutra A, Bouffard GG, Chang AM, Hansen NF, Wilfert AB, Thibaud-Nissen F, Schmitt AD, Belton J-M, Selvaraj S, Dennis MY, Soto DC, Sahasrabudhe R, Kaya G, Quick J, Loman NJ, Holmes N, Loose M, Surti U, Ra R, Graves Lindsay TA, Fulton R, Hall I, Paten B, Howe K, Timp W, Young A, Mullikin JC, Pevzner PA, Gerton JL, Sullivan BA, Eichler EE, Phillippy AM (2020) Telomere-to-telomere assembly of a complete human X chromosome. Nature 585:79–84
Milner SG, Jost M, Taketa S, Mazón ER, Himmelbach A, Oppermann M, Weise S, Knüpffer H, Basterrechea M, König P, Schüler D, Sharma R, Pasam RK, Rutten T, Guo G, Xu D, Zhang J, Herren G, Müller T, Krattinger SG, Keller B, Jiang Y, González MY, Zhao Y, Habekuß A, Färber S, Ordon F, Lange M, Börner A, Graner A, Reif JC, Scholz U, Mascher M, Stein N (2019) Genebank genomics highlights the diversity of a global barley collection. Nat Genet 51:319–326
Monat C, Padmarasu S, Lux T, Wicker T, Gundlach H, Himmelbach A, Ens J, Li C, Muehlbauer GJ, Schulman AH, Waugh R, Braumann I, Pozniak C, Scholz U, Mayer KFX, Spannagl M, Stein N, Mascher M (2019) TRITEX: chromosome-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol 20:284
Montenegro JD, Golicz AA, Bayer PE, Hurgobin B, Lee H, Chan C-KK, Visendi P, Lai K, Doležel J, Batley J, Edwards D (2017) The pangenome of hexaploid bread wheat. Plant J 90:1007–1013
Nilsen KT, Walkowiak S, Xiang D, Gao P, Quilichini TD, Willick IR, Byrns B, N’Diaye A, Ens J, Wiebe K (2020) Copy number variation of TdDof controls solid-stemmed architecture in wheat. Proc Natl Acad Sci 117:28708–28718
Niu S, Li J, Bo W, Yang W, Zuccolo A, Giacomello S, Chen X, Han F, Yang J, Song Y (2022) The Chinese pine genome and methylome unveil key features of conifer evolution. Cell 185(204–217):e214
Parra G, Blanco E, Guigó R (2000) GeneID in drosophila. Genome Res 10:511–515
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLOS Genetics 2:e190
Paux E, Sourdille P, Salse J, Saintenac C, Choulet F, Leroy P, Korol A, Michalak M, Kianian S, Spielmeyer W (2008) A physical map of the 1-gigabase bread wheat chromosome 3B. Science (New York, NY) 322:101–104
Pop M, Phillippy A, Delcher AL, Salzberg SL (2004) Comparative genome assembly. Brief Bioinform 5:237–248
Safár J, Simková H, Kubaláková M, Cíhalíková J, Suchánková P, Bartos J, Dolezel J (2010) Development of chromosome-specific BAC resources for genomics of bread wheat. Cytogenet Genome Res 129:211–223
Scalzitti N, Jeannin-Girardon A, Collet P, Poch O, Thompson JD (2020) A benchmark study of ab initio gene prediction methods in diverse eukaryotic organisms. BMC Genomics 21:293
Schreiber M, Mascher M, Wright J, Padmarasu S, Himmelbach A, Heavens D, Milne L, Clavijo BJ, Stein N, Waugh R (2020) A genome assembly of the barley ‘transformation reference’ cultivar golden promise. G3 Genes, Genomes, Genetics 10:1823–1827
Shang L, Li X, He H, Yuan Q, Song Y, Wei Z, Lin H, Hu M, Zhao F, Zhang C, Li Y, Gao H, Wang T, Liu X, Zhang H, Zhang Y, Cao S, Yu X, Zhang B, Zhang Y, Tan Y, Qin M, Ai C, Yang Y, Zhang B, Hu Z, Wang H, Lv Y, Wang Y, Ma J, Wang Q, Lu H, Wu Z, Liu S, Sun Z, Zhang H, Guo L, Li Z, Zhou Y, Li J, Zhu Z, Xiong G, Ruan J, Qian Q (2022) A super pan-genomic landscape of rice. Cell Res 32:878–896
Shi X, Cui F, Han X, He Y, Zhao L, Zhang N, Zhang H, Zhu H, Liu Z, Ma B, Zheng S, Zhang W, Liu J, Fan X, Si Y, Tian S, Niu J, Wu H, Liu X, Chen Z, Meng D, Wang X, Song L, Sun L, Han J, Zhao H, Ji J, Wang Z, He X, Li R, Chi X, Liang C, Niu B, Xiao J, Li J, Ling H-Q (2022) Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen-use efficiency in the wheat cultivar Kenong 9204. Mol Plant 15:1440–1456
Shiferaw B, Smale M, Braun H-J, Duveiller E, Reynolds M, Muricho G (2013) Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Security 5:291–317
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212
Soleimani B, Lehnert H, Keilwagen J, Plieske J, Ordon F, Naseri Rad S, Ganal M, Beier S, Perovic D (2020) Comparison between core set selection methods using different illumina marker platforms: a case study of assessment of diversity in wheat. Frontiers in Plant Science 11
Stanke M, Morgenstern B (2005) AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33:W465-467
Sun Y, Shang L, Zhu Q-H, Fan L, Guo L (2022) Twenty years of plant genome sequencing: achievements and challenges. Trends Plant Sci 27:391–401
Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203
Tao Y, Zhao X, Mace E, Henry R, Jordan D (2019) Exploring and exploiting pan-genomics for crop improvement. Mol Plant 12:156–169
van Berkum NL, Lieberman-Aiden E, Williams L, Imakaev M, Gnirke A, Mirny LA, Dekker J, Lander ES (2010) Hi-C: a method to study the three-dimensional architecture of genomes. Journal of Visualized Experiments: JoVE
Varshney RK, Nayak SN, May GD, Jackson SA (2009) Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol 27:522–530
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA (2001) The sequence of the human genome. Science (New York, NY) 291:1304–1351
Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez-Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature 588:277–283
Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, Mansueto L, Copetti D, Sanciangco M, Palis KC, Xu J, Sun C, Fu B, Zhang H, Gao Y, Zhao X, Shen F, Cui X, Yu H, Li Z, Chen M, Detras J, Zhou Y, Zhang X, Zhao Y, Kudrna D, Wang C, Li R, Jia B, Lu J, He X, Dong Z, Xu J, Li Y, Wang M, Shi J, Li J, Zhang D, Lee S, Hu W, Poliakov A, Dubchak I, Ulat VJ, Borja FN, Mendoza JR, Ali J, Li J, Gao Q, Niu Y, Yue Z, Naredo MEB, Talag J, Wang X, Li J, Fang X, Yin Y, Glaszmann J-C, Zhang J, Li J, Hamilton RS, Wing RA, Ruan J, Zhang G, Wei C, Alexandrov N, McNally KL, Li Z, Leung H (2018) Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557:43–49
Watson-Haigh NS, Suchecki R, Kalashyan E, Garcia M, Baumann U (2018) DAWN: a resource for yielding insights into the diversity among wheat genomes. BMC Genomics 19:941
Westman A, Kresovich S, Callow J, Ford-Lloyd B, Newbury J (1997) Biotechnology and plant genetic resources: conservation and use
Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Huang X, Li W, Li J, Liu Z, Li L, Liu J, Qi Q, Liu J, Li L, Li T, Wang X, Lu H, Wu T, Zhu M, Ni P, Han H, Dong W, Ren X, Feng X, Cui P, Li X, Wang H, Xu X, Zhai W, Xu Z, Zhang J, He S, Zhang J, Xu J, Zhang K, Zheng X, Dong J, Zeng W, Tao L, Ye J, Tan J, Ren X, Chen X, He J, Liu D, Tian W, Tian C, Xia H, Bao Q, Li G, Gao H, Cao T, Wang J, Zhao W, Li P, Chen W, Wang X, Zhang Y, Hu J, Wang J, Liu S, Yang J, Zhang G, Xiong Y, Li Z, Mao L, Zhou C, Zhu Z, Chen R, Hao B, Zheng W, Chen S, Guo W, Li G, Liu S, Tao M, Wang J, Zhu L, Yuan L, Yang H (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science (New York, NY) 296:79–92
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178:539–551
Zhao Q, Feng Q, Lu H, Li Y, Wang A, Tian Q, Zhan Q, Lu Y, Zhang L, Huang T, Wang Y, Fan D, Zhao Y, Wang Z, Zhou C, Chen J, Zhu C, Li W, Weng Q, Xu Q, Wang Z-X, Wei X, Han B, Huang X (2018) Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet 50:278–284
Zimin AV, Puiu D, Luo MC, Zhu T, Koren S, Marçais G, Yorke JA, Dvořák J, Salzberg SL (2017a) Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res 27:787–792
Zimin AV, Puiu D, Salzberg SL, Hall R, Kingan S, Clavijo BJ (2017b) The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. GigaScience 6
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2024 The Author(s)
About this chapter
Cite this chapter
N’Diaye, A., Walkowiak, S., Pozniak, C. (2024). Wheat Sequencing: The Pan-Genome and Opportunities for Accelerating Breeding. In: Appels, R., Eversole, K., Feuillet, C., Gallagher, D. (eds) The Wheat Genome. Compendium of Plant Genomes. Springer, Cham. https://doi.org/10.1007/978-3-031-38294-9_14
Download citation
DOI: https://doi.org/10.1007/978-3-031-38294-9_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-38292-5
Online ISBN: 978-3-031-38294-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)