Abstract
The evolutionary process of species is primarily determined by the variants within them, which are naturally selected according to the competitive advantage that these variants confer on the individuals who carry them. These changes or variants are called mutations, which can be gene mutations (also known as point mutations) or chromosomal mutations. Gene mutations alter one or a few nucleotides within the gene, while chromosomal ones affect entire portions of the chromosome (thus involving several genes) and even the entire chromosome. Chromosomal mutations can be of two types: structural and numerical. Structural chromosomal mutations involve a break in one or more points of one chromosome. If this breakage represents the loss of a section of the chromosome a deficiency occurs. If the segment that is lost on a chromosome is incorporated into its homologous chromosome, an addition takes place in the homologous chromosome. If the segment that breaks off is incorporated back into its original chromosome, but in an inverted form, an inversion occurs. If the segment that is lost is exchanged with a segment of another, non-homologous chromosome, it is a translocation. In numerical mutations, there are changes in the total number of chromosomes. These changes can be classified as cases of aneuploidy and cases of euploidy. In aneuploidy, there is the addition or loss of one or two chromosomes. In euploid individuals cells, there are a number of chromosomes that are a multiple of the basic genome of the species.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Author information
Authors and Affiliations
Solved Problems
Solved Problems
-
1.
A homozygous dominant plant for a certain trait (AA) is subjected to the action of ionizing radiation. This plant is used as a source of pollen to pollinate a homozygous recessive (aa) plant. From an offspring of 500 plants, two plants showed the phenotype corresponding to the recessive genotype. Can you explain this result?
With parents AA and aa, 100% of the offspring would be expected to have the phenotype determined by Aa. If aa plants appear, it is because some of the grains of pollen carried the “a” allele and not the “A” allele. Ionizing radiation may have been able to alter some DNA nucleotides, i.e., to have caused a point mutation, transforming the A allele into a.
-
2.
Explain how to identify cytologically through the microscope, the different structural chromosomal mutations.
During prophase I of meiosis, in the pachytene, the pairing of homologous chromosomes, in which each of the alleles of each gene of one of the homologous chromosomes will be exactly next to the corresponding alleles in the other homologous chromosome. If there is a change in the structure of one of the homologous chromosomes, the pairing of chromosomes will adopt characteristic shape to keep the alleles of the same gene side to side. Thus, if an insertion occurs, on the homologous chromosome of the chromosome with the insertion, the corresponding allele does not exist, so the segment of the DNA that has the insertion will not be able to pair, forming a loop (Fig. 10.4). In this figure, the chromosome of the right has an insertion, and the red loop represents the site of this chromosome where the insertion is. If there is a deficiency or deletion, there will likewise be a region of the chromosomes that do not have the same alleles, so a loop will form at the level of the chromosome site where the deficiency occurred, but on the normal homologous chromosome (Fig. 10.5). If an inversion occurs, the chromosome in which this mutation occurred will form a loop in order to mate with its normal counterpart (Fig. 10.6). In the case of translocations, the shape that paired chromosomes take is not predictable as in the cases of the above.
-
3.
A plant of chromosome number 2n = 2x = 10 crossed spontaneously with another plant of another species (2n = 4x = 20). The cross between these two plants produced mostly sterile offspring; however, some gametes were viable. F1 reductions were functional. Determine the number of chromosomes and the level of ploidy in the following cases:
-
(a)
F1
-
(b)
Backcrosses with plant 2n = 2x = 10
-
(c)
Backcrosses with plant 2n = 4x = 20
-
(d)
F1 chromosome duplication
-
(a)
-
(a)
Representing the chromosomes of the first plant as A (this does not indicate allelic information, indicates only that this is a chromosome of the first mentioned plant), and of the second plant as B, the gametes formed by them will be:
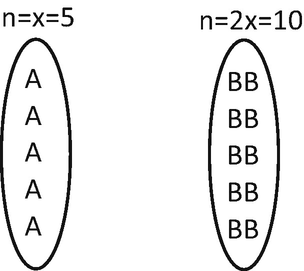
Therefore, the zygote of F1 will be:
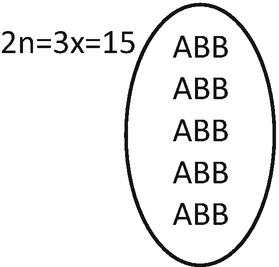
It will be an allotriploid individual (ABB), with 5 monovalent (A) and 5 bivalent (BB).
-
(b)
Because the functional gametes in F1 are non-reduced, their chromosome number is the same as that of F1 somatic cells (n = 3x = 15). When these gametes are merged with the gametes formed by the first mentioned plant (n = x = 5) is obtained:
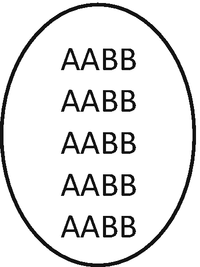
This individual will be allotetraploid (AABB), 2n = 4x = 20, with 10 bivalents (5 from one species and 5 from the other)
-
(c)
Merging of the gametes n = 2x = 10 and n = 3x = 15 will result in an allopentaploid individual (ABBBB) 2n = 5x = 25, with 5 monovalent and 5 tetravalent.
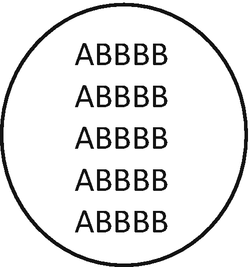
-
(d)
Chromosome duplication of F1 will result in a hexaploid (AABBBB) individual 2n = 6x = 30, with 5 bivalents (AA) and 5 tetravalent (BBBB).
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Laurentin Táriba, H.E. (2023). Chromosomal Mutations. In: Agricultural Genetics. Springer, Cham. https://doi.org/10.1007/978-3-031-37192-9_10
Download citation
DOI: https://doi.org/10.1007/978-3-031-37192-9_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-37191-2
Online ISBN: 978-3-031-37192-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)








