Abstract
Mammography is a popular diagnostic imaging procedure for detecting breast cancer at an early stage. Various deep-learning approaches to breast cancer detection incur high costs and are erroneous. Therefore, they are not reliable to be used by medical practitioners. Specifically, these approaches do not exploit complex texture patterns and interactions. These approaches warrant the need for labelled data to enable learning, limiting the scalability of these methods with insufficient labelled datasets. Further, these models lack generalisation capability to new-synthesised patterns/textures. To address these problems, in the first instance, we design a graph model to transform the mammogram images into a highly correlated multigraph that encodes rich structural relations and high-level texture features. Next, we integrate a pre-training self-supervised learning multigraph encoder (SSL-MG) to improve feature presentations, especially under limited labelled data constraints. Then, we design a semi-supervised mammogram multigraph convolution neural network downstream model (MMGCN) to perform multi-classifications of mammogram segments encoded in the multigraph nodes. Our proposed frameworks, SSL-MGCN and MMGCN, reduce the need for annotated data to 40% and 60%, respectively, in contrast to the conventional methods that require more than 80% of data to be labelled. Finally, we evaluate the classification performance of MMGCN independently and with integration with SSL-MG in a model called SSL-MMGCN over multi-training settings. Our evaluation results on DSSM, one of the recent public datasets, demonstrate the efficient learning performance of SSL-MNGCN and MMGCN with 0.97 and 0.98 AUC classification accuracy in contrast to the multitask deep graph (GCN) method Hao Du et al. (2021) with 0.81 AUC accuracy.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Graph modelling
- Self-supervised learning
- Semi-supervised learning
- Breast cancer classification
- Graph convolutional neural networks
1 Introduction
Breast cancer is the most common malignancy among adult women of all ages, accounting for over 7.8 million cases in the last five years [1]. Early detection of breast cancer improves survival rates by significantly limiting the risk of tumour progression and helping to increase patients’ life expectancy [2, 3]. Screening for cancers in mammography involves diagnosing methods to expose most breast malignancies in early stages. Radiologists diagnose these malignancies by detecting and examining the mass and calcification regions based on various visual signs, including size, edges, distribution, relations, and clustering [4, 5]. However, exposing these signs requires substantial expertise and are prone to high error rates of 20% [6]. Because of these challenges, and especially with the advancements in machine learning, recent years have witnessed dramatic developments of several computer vision models striving to extract enough hidden features from mammogram images to improve detection and classification sensitivity of breast cancers [7]. However, most of these techniques are significantly hindered by supervised machine learning approaches that require large datasets of accurately annotated images for training. Furthermore, in mammography, labelling malignancy regions, i.e., regions of interest (ROI), is a tedious procedure requiring pathologic expertise for considered time, making the process time-consuming and costly [8]. Thus, the availability of sufficiently labelled data is a critical bottleneck for supervised learning models, limiting the training, therefore, the performance and accuracy of the most recent models. As a result, current methods consistently adopt various techniques, including data augmentation, multi-view image generation, and transfer learning to mitigate inadequate data limitations and tune classification performance [9]. Work in [10] addressed the challenges of data limitation in the breast cancer domain by using transfer learning in CNN. The proposed method combined the pre-trained CNN, VGG16 [11], with a fully connected layer to perform binary classification of normal and abnormal mass in mammograms. Another work in [12] augmented the pre-trained VGG16 and Resnet50 [13] to a convolutional network model to perform a whole mammogram image classification. Authors in [14] applied multi-view, transfer learning and augmentation techniques to improve a CNN model performance with limited data.
Apparently, most of the techniques proposed to tackle the data limitation augmented to various end-to-end convolutional neural networks (CNNs) architectures, i.e., VGG16, Resnet, AlexNet, GoogleNet [15, 16]. CNNs employ fixed 2D kernels to encode images that contain well-defined and distinguishable objects, excluding the positions and orientations. However, mammography images are rich in heterogeneous textures that are difficult to classify based solely on their morphological shapes, so their geometric relations and dependencies should be considered [17].
Noticeably, a handful of approaches privilege the relationship between texture features to improve the performance of the CNN-based framework. Heyi Li et al. [33] augmented locality preserving and conditional graph learners module to a dual CNN model that maps between the ROIs and provided labels to improve the classification performance of breast mass. In addition, works in [25, 26] proposed a cross-view CNN model to construct the relationship between the features of two views of the mammograms, i.e., the mediolateral oblique (MLO) and the craniocaudal (CC). These techniques improve the performance of the mass detection models by exploiting the feature correlations. However, these methods lack generalisation capabilities as they are restricted to detecting the mass abnormalities in mammograms that are relatively large compared with other abnormalities such as calcifications clusters. More recently, graph-based deep learning approaches have demonstrated excellent advancements in machine learning, from solving complex geometric problems to handling massive data connections and learning data dependencies [18]. Moreover, relational awareness of graph-based models enables semi-supervised, and self-supervised learning approaches in various domains [20]. Consequently, graph-based models are proficient at circumventing the availability constraints of labelled mammograms by effectively privileging the inherited relations and dependencies in data to achieve improved accuracy with fewer labelled examples.
Very recently, several efforts have emerged to classify breast cancer using graphs, such as those used in [19, 21, 22]. These methods illustrate the advantages of graph-based models over conventional CNN models by modelling mammograms into graphs and performing binary graph classification. Another work in [17] highlights these advancements by performing a multi-classification of graphs modelled for calcification distributions in mammograms. The authors used the graph convolutional network (GCN) model that outperformed various CNN-based models, with a margin of over 10%. However, these techniques model ROIs in mammography into graphs, thus they are still limited because of the necessity of sufficiently well-annotated data.
Noticeably, in the entire cancer detection domain, significantly few graph-based models augment techniques to tackle the limitations of labelled data. For example, work in [23] proposed a weakly supervised GCN model to detect prostate cancer rates in histopathology slides. The proposed model outperforms the baseline supervised GCN by 36% and achieves 96% accuracy. Another method in [24] considers a self-supervised learning task to improve the performance of the graph neural networks (GNN) to classify breast cancer in histopathology images. The proposed approach outperforms other supervised GNN models by almost 20%. However, these methods assume a general classification of specific regions of histopathological images, which are less complex and computationally simple than mammography.
Considering all recent techniques, detecting and classifying breast cancer in mammography with minimal required annotated data and considering the relationship and pattern of the texture features is still an open problem. To the best of our knowledge, no self-supervised or semi-supervised graph-based technique has been previously proposed to process the high-resolution mammogram images and perform multi-classification of the anomalous regions with less annotated data requirements for the training process. However, as the learning capacities of graph-based models rely on the features and relations embedded in the graph, a well-engineered preprocess is necessary to transform the raw data of digitised mammogram images into a rich relational graph network.
This work models full mammogram images into efficient graph representations by capturing the heterogeneous features of high-level texture details and critical relations and patterns that contribute to diagnosing decisions. The proposed framework comprises a mammogram to multigraph transformer module (MMG) that segments the full-scale mammogram images into focused multi-region. It augments a pre-trained residual neural network (Res-Net) to transform each segment into high-level textures and spatial features called embeddings, resulting in a weighted graph. MMG also reinforces the features representation by generation multigraph that combines hundreds of graphs into a highly correlated network of thousands of nodes and edges.
The proposed framework includes a semi-supervised module, namely mammogram multigraph convolutional network module (dubbed MMGCN) for node classification. The MMGCN processes graph embeddings through stacked convolutional neural network layers followed by a fully connected network. It improves graph representations through semi-supervised learning replaces the embedding of each node with higher-level augmented embedding.
Furthermore, to reduce the need for a large annotated dataset, this work integrates a pre-training self-supervised learning process into the MMGCN by augmenting a self-supervised learning multigraph encoder (SSL-MG) to improve the feature representations. The SSL-MG improves the nodes embeddings through an adversarial process, discriminating between the series of node pairs, i.e., ordered and randomly generated nodes. Finally, the proposed framework classifies each node into normal cells or any of the breast abnormalities, i.e., mass malignant or benign and calcification malignant or benign.
2 Proposed Method
2.1 Notations and Problem Definition
Given a mammogram dataset D that consists of a number of images, \(I=\left\{ I_{i}\right\} _{1}^{|D|}\). Let each image \(I_{i}\) can be divided into K segments \(S=\left\{ S_{i}\right\} _{1}^{|K|}\) where each segment \(S_{i}\) has texture features \(S_{i}^{T}\), spatial details \(S_{i}^{S}\), and category \(\quad S_{i}^{C} \in \{0:normal, 1:massMalignant, 2:massBenign, 3:calcificationMalignant, 4:calcificationBenign\}\).
Each image \(I_{i}\) can be modelled as a graph \(G_{i}=(V, E)\) where \(V \le |S|\) is the set of nodes assigned to non-zero segments, and \(E \subseteq A=V \times V\) is a set of edges connecting the nodes based on an adjacency matrix A. If \(\left\{ v_{i}, v_{j}\right\} \in \ V\) are two nodes representing adjacent segments, so an edge connect them and denoted as \(e_{i,j}\in \ E\). Graph \(G_i\) is weighted using the correlation between the segmented images as features \({H_{E}}\) added to all edges E and the vectorization of the high level texture features of the image segments S as features \({H_{V}}\in \mathbb {R}^{d|S|}\) added to all nodes.
Modelling a complete mammogram dataset D consisting of |D| images generating a set of weighted graphs \(G=\left\{ {G}_{i}\right\} _{1}^{|D|}\). In order to enrich the encoded mammograms features and relationships, a complex multi-graph \(\mathcal {G}\) is constructed by connecting all graphs as a united graph \(\mathcal {G}=\bigcup _{G_{1}}^{G_{|D|}}{(G_i)}\).
Given a multi-graph \(\mathcal {G}\) with initial embeddings \({H}^{0}\) and a small subset of labelled nodes \(V^L\), our aim in this work is to improve graph representation through a self-supervised pretext task. Then use semi-supervised downstream model to computes the loss between the given labels \(S^{C}\) and embeddings \(H^{l}\) of labelled nodes \(V^L\) and update the learnable weight W. Finally each node gets final embedding \({Z}_{i}\) and as each \({Z}_{i}\) present a segment in a mammogram, so each segment get classified as \(S_{i}{\rightarrow }S_{i}^{C}\) with better accuracy than predicting a general class for the whole image.
2.2 Mammograms to Multi-Graph Modelling (MMG)
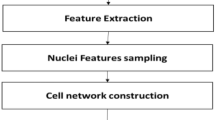
This work proposes a mammogram multi-graph transformer (MMG) as presented in Fig. 1 and given in Algorithm 1 in the appendix. Mammograms are high-resolution images composed of heterogeneous pixels with values varying between black and white, i.e., \(0 \sim 1 \). To fully capture the features in these mammograms, the proposed MMG module transforms each image to a graph embedded with texture and spatial features representing nodes and edges.
Initially, MMG divides each mammogram image into K segments, then encode the texture features \(S_{i}^{T}\) of these segments using a pre-trained ResNet-18 [27] model. ResNet-18 is composed of a series of residual blocks of localised convolutional and pooling layers that vectorise the texture features \(S_{i}^{T}\) of each sub-image \(S_i\) into a 512-length vector \(\overrightarrow{\boldsymbol{X}}\) as given in Eq. (1). MMG embeds the encoded vectors \(\overrightarrow{\boldsymbol{X}}\) as node features \({H_{V}}\) embedded into graph nodes V.
Here \(S_{i}^{T}\) and \(\overrightarrow{\boldsymbol{X}}\) denotes input and output of the residual network layers, while \(W_{i}\) and \(W_{s}\) represents the layers and linear projections of the ResNet.
MMG encodes the Cartesian coordinates of each mammogram segments to generate edges list and a adjacency matrix A defining the connected nodes in each graph \(\mathcal {G}_{i}\). In order to preserve the correlation between the segmented images of the mammogram, MMG uses the cosine similarity [28] illustrated in Eq.(2) to weight graph edges by values varying between 1 for edges connecting nodes representing the same features and 0 for pairs of nodes with entirely unmatched features.
\(A_{k}\) and \(B_{k}\) denote vectors A and B components, whereas n represents the number of components. As equal length N-dimensional arrays represent both vectors, the components are the elements of these arrays. MMG optimises the generated graph by pruning nodes and edges representing the background segments of the mammogram image. Then it assigns a class for each node using the region of interest (ROI) binary masks. The binary mask consists of pixels with 0 values except for the region of abnormality with pixel values of 1. MMG combines the optimised graphs using the common nodes in non-Euclidean spaces to generate the final complex multi-graph network as given in Eq. (3). The equation unites a set of N graphs representing the entire mammography dataset images D where \(N=|D|\) and each graph is composed of nodes \(\mathcal {V}\), edges \(\mathcal {E}\), and features \(\mathcal {H_{V}, H_{E}}\).
Now \(\mathcal {G}\) is the modelled graph network for the entire mammogram dataset. As \(\mathcal {G}\) composes all segments of mammogram images as nodes, each node can be classified based on the embedded features and the relation to other nodes into one of 5 classes. These classes are normal, mass-Malignant, mass-Benign, calcification-Malignant, and calcification-Benign.
2.3 Multi-Graph Self-Supervised Learning (SSL-MG)
This stage process the modelled mammogram multi-graph \(\mathcal {G}\) by the proposed SSL-MG encoder to improves the segmented image features embedded in nodes based on a self-supervised pretext task. SSL-MG encoder comprises nodes and graph readers, discriminators, and GCN layers [32] stacked with pooling and fully connected layers. SSL-MG employs a mini-batch generator [29] to process the multi-graph \(\mathcal {G}\) as a series of sub-graphs \(\mathcal {G^{\star }}\) to fit less memory. As features of the mammogram segmented images are vectors \(H_V\) of length K embedded in multi-graph nodes have large scale varies values, so SSL-MG normalises them for better computation to values between 0 and 1 using Eq. (4).
Additionally, the weighted adjacency matrix A of the mammogram multi-graph is normalised using the symmetric normalisation trick illustrated by Kipf and Welling in [20]. Equation (5) normalises A after adding self connection for all nodes using the unit matrix \(I_{N}\) then multiples it with the two inverses of the square root of the degree matrix D [32].
SSL-MG first aggregates and down-samples the features H into an embedding \(Z^\star \) that summarises the sub-graph \(\mathcal {G^{\star }}\). Equation (6) computes \(Z^\star \) by matrix multiplication of the normalised adjacency matrix of the sub-graph \(\widehat{\textrm{A}}^\star \), the normalised features \(\widehat{\textrm{H}}\), and network weight W. SSL-MG then uses this embedding in a self-supervised pretext task to discriminate between a series of features, one for the nodes of the same sub-graph \(h_i\) and another for random nodes \({h}_{i}^{T}\) (Fig. 2).
SSL-MG encoder: The encoder processes the complex multi-graph network in batches, generates a graph summary of each batch, and compares it to the embeddings of pair of nodes, one in sorted order and the second using random node. The loss function at the end compares the similarity of these embeddings.
SSL-MG process three inputs include embeddings of the sorted nodes \({h_i}\), embeddings of an opposing random node \(h^{T}_{i}\), and the computed graph summary \({Z}^\star \). The encoder learns the node presentation by maximizing the similarities between the sorted nodes and the graph summary while decreasing it for the random nodes. For that, SSL-MG in Eq. (7) uses logistic sigmoid non-linear function \(\sigma \) to compute the probability of \(\left( h_i,{{Z}^\star }\right) \) and \(\left( h^{T}_{i},{{Z}^\star }\right) \), then compute the sub-graph sigmoid cross-entropy loss \(\mathcal {L^\star }_{S C E}\) for all the nodes M and N. The total loss \(\mathcal {L}_{S C E}\) then calculated by aggregating the loss of a k of \(\mathcal {G}^\star \)
\(SSL-MG\) encoder minimizes the cross-entropy loss calculated by Eq. (7) by using the adaptive momentum (Adam) function. This let the encoder learn the representation of the graph and generate high-level embeddings to replace the existing for each node. The MG-SSL encoder tunes the features of segmented mammogram images embedded in the multi-graph nodes. Later, these embeddings are used as an input for the downstream model.
2.4 Mammogram Multi-Graph Convolutional Network Classifier (MMGCN)
MMGCN is a multi-node classifier model designed to either processes the initial features of the mammogram segmented images embedded in the multi-graph or the tuned nodes embeddings generated from the SSL-MG encoder as depicted in Fig. 3. MMGCN processes the input of the mammogram multi-graph batches \(\mathcal {G^{\star }}\) same way as the SSL-MG by normalising the nodes embeddings and adjacency matrix using Eqs. (4) and (5) respectively. In addition, MMGCN employs a data balancing procedure to guarantee that the nodes categories \(\quad S_{i}^{C} \in \{0:normal, 1:massMalignant, 2:massBenign, 3:calcificationMalignant, 4:calcificationBenign\}\) are presented equally in each sub-graph \(\mathcal {G^{\star }}\). As the mammogram multi-graph includes nodes represent images segments of normal sections in large numbers compared to the other categories, this step required to avoid any bias through the downstream process.
MMGCN includes 4 GCN layers to aggregate the features of each node and its neighbours, then normalises and processes each aggregation with learnable weight W through a standard dense layer. The GCN layers perform that through matrix multiplication of the normalised adjacency matrix with self-connection \(\widehat{\mathcal {A}}\star \), the normalised features matrix \(\widehat{\mathcal {X}}\) and the learnable weight W. As in Eq. 8, these multiplications get activated using none linear function typically Relu. However, the last GCN layer uses softmax activation function as in Eq. 9.
As the initial mammogram multi-graph composes nodes, each one is embedded with the encoded features \(h_i\) of a single image segment \(S_i\). Now MMGCN generates higher-level embedding \(Z_i\) that embeds features of all neighbour segments in each node. By getting an embedding \(Z_{i}=\sum _{i} \exp \left( h_{V_{i}}\right) \) for each node, the softmax uses it to calculate the probability of each node class \(p\left( \mathcal {S}_{i}^{C}\mid \textbf{Z}_{i}\right) \).
The MMGCN model either processes the initial mammogram multi-graph generated by the MMG or the improved multi=graph generated by SSL-MG encoder. The MMGCN use a mini-batch generator and sparse convolution layer as an input layer to process all the input tensors efficiently. Additionally, the model comprises of four graph convolutional layers of 512, 128, 64, and 32 units, followed by an aggregation of indices and a fully connected layer to sort and compute the output followed by softmax function.
Using a subset of labelled nodes \(V^{L} \in \ V\) represent annotated mammogram segments \(S_{i}^{C}\in \ S\), the categorical cross-entropy loss \(\mathcal {L}_{C C E}\) can be calculated through a semi-supervised training using Eq. (10). Finally, a stochastic gradient descent optimiser uses this loss to train the neural network weights W.
3 Experiments
3.1 Dataset
We validate our frameworks, i.e., MMGCN, and SSL-MMGCN, with public mammography dataset, CBIS-DDSM [31]. The dataset contains scanned images of digitised mammograms in the digital imaging and communications in medicine format (DICOM), a standard format for screening in the medical domain. The dataset contains 2,620 mammography images in two standard views, MLO and CC. In addition, CBIS-DDSM has training samples that include annotation binary masks for the ROI that indicate the general positions of anomalies within mammograms. The dataset included 557 patient mammograms with calcification anomalies, 646 with mass anomalies, and 45 with both anomalies. Moreover, each type of anomaly is classified as either malignant or benign. The mammograms in the raw data have varied large-scale dimensions to provide enough capability for zooming and analysis. Using the CBIS-DDSM dataset, MMG encodes 1138 mammograms in a complex multi-graph. This multi-graph contains 285413 nodes: 3478 represent mass-malignant regions, 2928 represent mass-benign regions, 1596 represent calcification-malignant regions, and 2033 represent calcification-benign regions, while the remaining nodes encode normal lesions.
3.2 Experiment Setup
The experiment setup is crucial in machine learning, as we should consider various measurements to avoid data leakage, overfitting, and bias. Especially in graph learning message passing and feature smoothing over neighbouring nodes. Hence, for the training process of the SSL-MMGCN and MMGCN models, we load a multi-graph with 40% and 50% of labelled nodes from each class, respectively. We include the remaining nodes unlabeled in the multi-graph for the validation process. Then, to avoid bias during the training process, the node balancing module generates an equal number of nodes from each class in each mini-batch. Also, the MMGCN model employs a 0.5 drop rate to reduce overfitting and perform smooth learning.
3.3 Performance Evaluation
SSL-MMGCN Learning. The SSL-MG encoder is trained on a multi-graph network of 7500 unlabelled nodes for training over 200 epochs. The convergence of the model optimiser is depicted in Fig. 4(a). Over the first 50 epochs, the decay rate demonstrates a rapid convergence with a drop in the loss value from 0.7 to less than 0.1. However, with further training over the last 150 epochs, the loss steadily declines to a value close to zero. Through the training process of the SSL-MG, the model learns the node and graph representation and replaces the features of the nodes with higher-level information based on the learning efficiency of the self-supervised task. Then, by using the generated embedding as an input for the MMGCN in the SSL-MMGCN framework, semi-supervised learning training is performed using only 50% of the nodes, while the rest are for validation and testing. The SSL-MMGCN training and validation loss rate over 1000 training epochs illustrated in Fig. 4(b) shows a decrease to less than 0.25. The decline in losses and the modest variations between the training and validation losses indicate that the downstream SSL-MMGCN model has an effective learning rate. Figure 4(c) shows the accuracy improvement of the SSL-MMGCN model through this training. The model accuracy efficiently exceeds 95% at the end of the 1000 training epochs with a continuous learning rate, albeit a slow learning rate after 900 epochs, which implies the convergence of the SSL-MMGCN model. The significant increase in training and validation accuracy rates shows the learning capacity’s efficacy, especially with the labelled to unlabelled data ratio. SSL-MMGCN uses only 50% of the multi-graph nodes to calculate the categorical cross-entropy loss and adjust the learnable weight W using the ADAM optimizer. The loss in Fig. 4(b) and the accuracy in Fig. 4(c) demonstrate continuous gradient descent, learning without over-fitting. However, after 300 epochs, the loss increases and the accuracy decreases, which illustrates a non-optimal local minimum. However, after a few epochs, the model optimises with better gradient descents. The efficient fitting of the model shows that increasing the number of labelled nodes or training epochs allows SSL-MMGCN to attain improved accuracy.
Mammogram Classification Analysis. In the medical domain, confusion among the classes is crucial in the diagnosis process, and the percentage of false and true positives is considered. So, to investigate the sensitivity and specificity of the MMGCN model in classifying the categories of breast cancer anomalies in mammography, the confusion matrix is computed, as shown in Fig. 6. The results show that the true-positive classification of the MMGCN across all categories varies between 97.33% and 99.13%. The maximum confusion is for classifying calcification-malignancy, where 1% is wrongly classified as benign and less than 2% among the mass and normal classes. The mass malignant and calcification benign have the same confusion rates, while the minimum confusion rate is less than 1% for classifying the normal segments wrongly.
To analyse the ability of SSL-MGCN to distinguish between the five classes, we use the ROC curve evaluation metric. This curve plots the probability of each class’s true-positive versus false-positive rates, considering one-to-all classes. Figure 5 shows the ROC curves of all five classes, which demonstrate the model’s effectiveness in classifying each class correctly with almost 100%, albeit the model can misclassify the malignant calcification anomalies by 1%.
3.4 Compared Methods
To demonstrate the advantage of modelling the segments of the mammogram images into a multigraph and integrating a self-supervised pre-training encoder, we compare our frameworks, i.e., MMGCN and SSL-MMGCN, to the current state-of-the-art methods in [33, 34, 36, 37]. Table 1 lists the performance of each method as presented in their papers, including the AUC accuracy, the considered abnormalities, and the classification task. Furthermore, for fair analyses, we consider the train-test ratio for each experiment setup.
Similar to our framework, only the work in [37] adopted a whole mammogram multi-classification method to detect both the calcification and mass abnormalities. Further, the other methods are limited to the classification of only one type of abnormality, the mass abnormalities as in [33] and the calcification abnormalities as in [34].
Compared to our framework, which enhances the graph embedding by integrating a self-supervised encoder and reduces the learning rates by adopting a semi-supervised graph-based model, other methods only integrate fully supervised methods. As a result, our method requires less annotated data for training, 40% for SSL-MMGCN, and 60% for MMGCN, compared to 80% in the other methods. However, MMGCN and SSL-MMGCN outperform these methods, particularly the framework proposed in [33] and [37], which use the same dataset, i.e., DDSM, for evaluation.
Noticeably, the work in [33] has better AUC accuracy when evaluated on the INbreast dataset rather than the DDSM dataset we use in our experiments. That is because of the better resolution quality of the full-field digital mammogram images in INbreast than the digitised images in DDSM. So, in extended experiments, we will evaluate our framework on the most recent digital mammogram dataset, which can result in even better AUC accuracy.
4 Conclusion
This work adopts a graph-based deep learning framework that enables semi-supervised and self-supervised machine learning approaches to perform efficient breast cancer classification using mammogram data. The framework models the heterogeneous high-level texture features and their critical relations and spatial details inherent to mammograms. MMG maps each mammogram to a graph and later combines these graphs into a multi-graph to improve the representation of the relations and features in a mammogram. To perform node-level classification, we have exploited the benefits of MMGCN and SSL-MMGCN models where pre-trained self-supervised SSL-MMGCN demonstrates significant improvement in learning with limited labeled data. Self-supervision significantly improves the training time in the downstream process. Results show that with sufficient labeled data, i.e., 40% or more, the MMGCN model shows accelerated learning capacity and better multi-classification sensitivity.
Experiments results reveal the proposed graph-based framework has excellent AUC classification performance of 0.97 for the SSL-MMGCN and 0.98 for the MMGCN and outperforms state-of-the-art works for breast cancer diagnosis, including Li H. et al. [33], Hao Du et al. [34]and Le et al. [37].
In future works, we will consider the augmentation of other convolutional neural networks to encode mammogram features efficiently to accelerate accurate breast cancer diagnosis with possible consideration in clinical trials.
References
Bataille, V., et al.: Nevus size and number are associated with telomere length and represent potential markers of a decreased senescence in vivo. Cancer Epidemiol. Prev. Biomark. 16(7), 1499–1502 (2007)
Kösters, J.P., Gøtzsche, P.C.: Regular self-examination or clinical examination for early detection of breast cancer. Cochrane Database Syst. Rev. (2) (2003)
Mordang, J.J., et al.: The importance of early detection of calcifications associated with breast cancer in screening. Breast Cancer Res. Treat. 167(2), 451–458 (2018)
Hofvind, S., Iversen, B.F., Eriksen, L., Styr, B.M., Kjellevold, K., Kurz, K.D.: Mammographic morphology and distribution of calcifications in ductal carcinoma in situ diagnosed in organized screening. Acta Radiol. 52(5), 481–487 (2011)
Nalawade, Y.V.: Evaluation of breast calcifications. Indian J. Radiol. Imaging 19(4), 282–286 (2009)
Bejnordi, B.E., et al.: Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318(22), 2199–2210 (2017)
Henriksen, E.L., Carlsen, J.F., Vejborg, I.M., Nielsen, M.B., Lauridsen, C.A.: The efficacy of using computer-aided detection (CAD) for detection of breast cancer in mammography screening: a systematic review. Acta Radiologica 60(1), 13–18 (2019)
Katalinic, A., Bartel, C., Raspe, H., Schreer, I.: Beyond mammography screening: quality assurance in breast cancer diagnosis (The QuaMaDi Project). Br. J. Cancer 96(1), 157–161 (2007)
Abdelhafiz, D., Yang, C., Ammar, R., Nabavi, S.: Deep convolutional neural networks for mammography: advances, challenges and applications. BMC Bioinform. 20(11), 1–20 (2019)
Guan, S., Loew, M.: Breast cancer detection using transfer learning in convolutional neural networks. In: Conference on AIPR 2017 IEEE Applied Imagery Pattern Recognition Workshop, pp. 1–8 (2017)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint (2014). arXiv:1409.1556
Shen, L., Margolies, L.R., Rothstein, J.H., Fluder, E., McBride, R., Sieh, W.: Deep learning to improve breast cancer detection on screening mammography. Sci. Rep. 9(1), 1–12 (2019)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Salama, W.M., Wessam, M., Aly, M.H.: Deep learning in mammography images segmentation and classification: automated CNN approach. Alex. Eng. J. 60(5), 4701–4709 (2021)
Ballester, P., Araujo, R.M.: On the performance of GoogLeNet and AlexNet applied to sketches. In: Thirtieth AAAI Conference on Artificial Intelligence (2016)
Alom, M.Z., et al.: The history began from alexnet: a comprehensive survey on deep learning approaches. arXiv preprint (2018). arXiv:1803.01164
Du, H., Yao, M.M.S., Chen, L., Chan, W.P., Feng, M.: Multi-task Graph Convolutional Neural Network for Calcification Morphology and Distribution Analysis in Mammograms. arXiv preprint (2021). arXiv:2105.06822
Zhang, Z., Lee, W.S.: Deep graphical feature learning for the feature matching problem. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 5087–5096 (2019)
Gallego-Ortiz, C., Martel, A.L.: A graph-based lesion characterization and deep embedding approach for improved computer-aided diagnosis of nonmass breast MRI lesions. Med. Image Anal. 51, 116–124 (2019)
Kipf, T.N., Thomas, N., Welling, M.: Semi-supervised classification with graph convolutional networks. arXiv preprint (2016). arXiv:1609.02907
Du, H., Feng, J., Feng, M.: Zoom in to where it matters: a hierarchical graph based model for mammogram analysis. arXiv preprint (2019). arXiv:1912.07517
Zhang, Y.D., Satapathy, S.C., Guttery, D.S., Górriz, J.M., Wang, S.H.: Improved breast cancer classification through combining graph convolutional network and convolutional neural network. Inf. Process. Manag. 58(2), 102439 (2021)
Wang, J., Chen, R.J., Lu, M.Y., Baras, A., Mahmood, F.: Weakly supervised prostate TMA classification via graph convolutional networks. In: Conference on ISBI 2020 IEEE 17th International Symposium on Biomedical Imaging, pp. 239–243 (2020)
Özen, Y.: Self-supervised representation learning with graph neural networks for region of interest analysis in breast histopathology. Doctoral dissertation, Bilkent University (2020)
Ma, J., Li, X., Li, H., Wang, R., Menze, B., Zheng, W.S.: Cross-view relation networks for mammogram mass detection. In: Conference on ICPR 2020 25th International Conference on Pattern Recognition, pp. 8632–8638 (2021)
Yang, Z., et al.: MommiNet-v2: mammographic multi-view mass identification networks. Med. Image Anal. 73, 102204 (2021)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
Dehak, N., Dehak, R., Glass, J.R., Reynolds, D.A., Kenny, P.: Cosine similarity scoring without score normalization techniques. In: Odyssey, p. 15 (2010)
Mondal, A.K., Jain, V., Siddiqi, K.: Mini-batch graphs for robust image classification. arXiv preprint (2021). arXiv:2105.03237
Van der Maaten, L., Hinton, G.: Visualizing data using t-SNE. J. Mach. Learn. Res. 9(11) (2008)
Lee, R.S., Gimenez, F., Hoogi, A., Miyake, K.K., Gorovoy, M., Rubin, D.L.: A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 4(1), 1–9 (2017)
Kipf, T.N., Welling, M.: Semi-supervised classification with graph convolutional networks. arXiv preprint (2016). arXiv:1609.02907
Li, H., Chen, D., Nailon, W.H., Davies, M.E., Laurenson, D.I.: Dual convolutional neural networks for breast mass segmentation and diagnosis in mammography. IEEE Trans. Med. Imaging 41(1), 3–13 (2021)
Du, H., Yao, M.M.S., Chen, L., Chan, W.P., Feng, M.: Multi-task Graph Convolutional Neural Network for Calcification Morphology and Distribution Analysis in Mammograms. arXiv preprint, vol. 14 (2021). arXiv:2105.06822
Dhungel, N., Carneiro, G., Bradley, A.P.: The automated learning of deep features for breast mass classification from mammograms. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 106–114. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_13
Al-Antari, M.A., Al-Masni, M.A., Kim, T.S.: Deep learning computer-aided diagnosis for breast lesion in digital mammogram. Deep Learn. Med. Image Anal. 59–72 (2020)
Le, T.L.T., Thome, N., Bernard, S., Bismuth, V., Patoureaux, F.: Multitask classification and segmentation for cancer diagnosis in mammography. arXiv preprint (2019). arXiv:1909.05397
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2023 The Author(s)
About this paper
Cite this paper
Ibrahim, M., Henna, S., Cullen, G. (2023). Multi-Graph Convolutional Neural Network for Breast Cancer Multi-task Classification. In: Longo, L., O’Reilly, R. (eds) Artificial Intelligence and Cognitive Science. AICS 2022. Communications in Computer and Information Science, vol 1662. Springer, Cham. https://doi.org/10.1007/978-3-031-26438-2_4
Download citation
DOI: https://doi.org/10.1007/978-3-031-26438-2_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-26437-5
Online ISBN: 978-3-031-26438-2
eBook Packages: Computer ScienceComputer Science (R0)