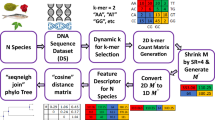
Abstract
Top-k similar sequence search is an essential tool for DNA data management. Given a DNA database, it is a problem to extract k similar DNA sequence pairs in the database, which yield the highest similarity among all possible pairs. Although this is a fundamental problem used in the bioinformatics field, it suffers from an expensive computational cost. To overcome these limitations, we propose a novel fast top-k similarity search algorithm for DNA databases. We conducted experiments using real-world DNA sequence datasets, and experimentally confirmed that the proposed method achieves a faster top-k search than baseline algorithms while keeping high accuracy.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Li, G., Deng, D., Wang, J., Feng, J.: PASS-JOIN: a partition-based method for similarity joins. Proc. VLDB Endow. 5(3), 253–264 (2011)
Wang, W., Qin, J., Xiao, C., Lin, X., Shen, H.T.: VChunkJoin: an efficient algorithm for edit similarity joins. IEEE Trans. Knowl. Data Eng. 25(8), 1916–1929 (2013)
Zhang, H., Zhang, Q.: EmbedJoin: efficient edit similarity joins via embeddings. In: Proceedings of KDD (2017)
Zhang, H., Zhang, Q.: MinJoin: efficient edit similarity joins via local hash minima. In: Proceedings of KDD (2019)
Suzuki, Y., Sato, M., Shiokawa, H., Yanagisawa, M., Kitagawa, H.: MASC: automatic sleep stage classification based on brain and myoelectric signals. In Proceedings of ICDE (2017)
Wagner, R.A., Fischer, M.J.: The string-to-string correction problem. J. ACM 21, 168–173 (1974)
Shiokawa, H.: Scalable affinity propagation for massive datasets. In Proceedings of AAAI (2021)
Deng, D., Li, G., Feng, J., Li, W.-S.: Top-k string similarity search with edit-distance constraints. In: Proceedings of ICDE (2013)
Yang, Z., Jianjun, Yu., Kitsuregawa, M.: Fast algorithms for top-k approximate string matching. In Proceedings of AAAI (2010)
Yangjun, C., Nguyen, H-H.: On the string matching with k differences in DNA databases. Proc. VLDB 14(6), 903–915 (2021)
Yangjun, C., Yujia, W.: On the string matching with k mismatches. Theoret. Comput. Sci. 726 (2018). https://doi.org/10.1016/j.tcs.2018.02.001
Yangjun, C., Yujia, W.: BWT arrays and mismatching trees: a new way for string matching with k mismatches. In: Proceedings of ICDE (2017)
Ukkonen, E.: Algorithms for approximate string matching. Inf. Control 64, 100–118 (1985)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Yagi, R., Shiokawa, H. (2022). Fast Top-k Similar Sequence Search on DNA Databases. In: Pardede, E., Delir Haghighi, P., Khalil, I., Kotsis, G. (eds) Information Integration and Web Intelligence. iiWAS 2022. Lecture Notes in Computer Science, vol 13635. Springer, Cham. https://doi.org/10.1007/978-3-031-21047-1_14
Download citation
DOI: https://doi.org/10.1007/978-3-031-21047-1_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-21046-4
Online ISBN: 978-3-031-21047-1
eBook Packages: Computer ScienceComputer Science (R0)