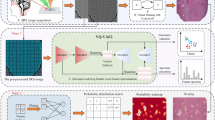
Abstract
Predicting complete response to radiotherapy in rectal cancer patients using deep learning approaches from morphological features extracted from histology biopsies provides a quick, low-cost and effective way to assist clinical decision making. We propose adjustments to the Vision Transformer (ViT) network to improve the utilisation of contextual information present in whole slide images (WSIs). Firstly, our position restoration embedding (PRE) preserves the spatial relationship between tissue patches, using their original positions on a WSI. Secondly, a clustering analysis of extracted tissue features explores morphological motifs which capture fundamental biological processes found in the tumour micro-environment. This is introduced into the ViT network in the form of a cluster label token, helping the model to differentiate between tissue types. The proposed methods are demonstrated on two large independent rectal cancer datasets of patients selectively treated with radiotherapy and capecitabine in two UK clinical trials. Experiments demonstrate that both models, PREViT and ClusterViT, show improvements in the prediction over baseline models.
Supported by Cancer Research UK.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Andréé, T., Shiu, K., Kim, T., et al.: Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med. 383(23), 2207–2218 (2020). https://doi.org/10.1056/NEJMoa2017699
Anitei, M.G., et al.: Prognostic and predictive values of the immunoscore in patients with rectal cancer. Clin. Cancer Res. 20(7), 1891–1899 (2014). https://doi.org/10.1158/1078-0432.CCR-13-2830
Bilal, M., Raza, S., Azam, A., et al.: Development and validation of a weakly supervised deep learning framework to predict the status of molecular pathways and key mutations in colorectal cancer from routine histology images: a retrospective study. Lancet Digit Health 3(12), e763–e772 (2021)
Bychkov, D., Linder, N., Turkki, R., et al.: Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 8, 3395 (2018). https://doi.org/10.1038/s41598-018-21758-3
Campanella, G., Hanna, M., Geneslaw, L., et al.: Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25, 1301–1309 (2019). https://doi.org/10.1038/s41591-019-0508-1
Dosovitskiy, A., et al.: An image is worth 16 \(times\) 16 words: transformers for image recognition at scale. CoRR (2020). arxiv.org/abs/2010.11929
Echle, A., Grabsch, H.I., Quirke, P., et al.: Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology 159(4), 1406–1416 (2020)
Gao, Z., et al.: Instance-based vision transformer for subtyping of papillary renal cell carcinoma in histopathological image. CoRR (2021). arxiv.org/abs/2106.12265
George, T.J.J., Allegra, C.J., Yothers, G.: Neoadjuvant rectal (NAR) score: a new surrogate endpoint in rectal cancer clinical trials. Curr. Colorectal Cancer Rep. 11(5), 275–280 (2015). https://doi.org/10.1007/s11888-015-0285-2
Guinney, J., Dienstmann, R., Wang, X., et al.: The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015). https://doi.org/10.1038/nm.3967
Hildebrand, L.A., Pierce, C.J., Dennis, M., Paracha, M., Maoz, A.: Artificial intelligence for histology-based detection of microsatellite instability and prediction of response to immunotherapy in colorectal cancer. Cancers (Basel) 13(3), 391 (2021). https://doi.org/10.3390/cancers13030391
Iizuka, O., Kanavati, F., Kato, K., et al.: Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci. Rep. 10, 1504 (2020). https://doi.org/10.1038/s41598-020-58467-9
Ilse, M., Tomczak, J., Welling, M.: Attention-based deep multiple instance learning. In: Dy, J., Krause, A. (eds.) Proceedings of the 35th International Conference on Machine Learning. Proceedings of Machine Learning Research, vol. 80, pp. 2127–2136. PMLR (2018). https://proceedings.mlr.press/v80/ilse18a.html
Islam, M.A., Jia, S., Bruce, N.D.B.: How much position information do convolutional neural networks encode? CoRR (2020). arxiv.org/abs/2001.08248
Jones, H.J.S., et al.: Stromal composition predicts recurrence of early rectal cancer after local excision. Histopathology 79, 947–956 (2021). https://doi.org/10.1111/his.14438
Kanavati, F., Toyokawa, G., Momosaki, S., et al.: A deep learning model for the classification of indeterminate lung carcinoma in biopsy whole slide images. Sci. Rep. 11, 8110 (2021). https://doi.org/10.1038/s41598-021-87644-7
Kim, N., et al.: Detection of microsatellite instability in colorectal cancer patients with a plasma-based real-time PCR analysis. Front. Pharmacol. 12 (2021). https://doi.org/10.3389/fphar.2021.758830,www.frontiersin.org/article/10.3389/fphar.2021.758830
Koelzer, V., Lugli, A., Dawson, H., et al.: Cd8/cd45ro t-cell infiltration in endoscopic biopsies of colorectal cancer predicts nodal metastasis and survival. J. Trans. Med. 12(81) (2014). https://doi.org/10.1186/1479-5876-12-81
Li, H., et al.: DT-MIL: deformable transformer for multi-instance learning on histopathological image. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12908, pp. 206–216. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87237-3_20
Ling, C.X., Huang, J., Zhang, H.: AUC: a better measure than accuracy in comparing learning algorithms. In: Xiang, Y., Chaib-draa, B. (eds.) Advances in Artificial Intelligence, pp. 329–341. Springer, Berlin Heidelberg (2003). https://doi.org/10.1007/3-540-44886-1_25
Lu, M.Y., Williamson, D., Chen, T.Y., Chen, R.J., Barbieri, M., Mahmood, F.: Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5(6), 555–570 (2021). https://doi.org/10.1038/s41551-020-00682-w
New treatment could spare early-stage rectal cancer patients life-altering side effects. www.birmingham.ac.uk/news/2020/new-treatment-could-spare-early-stage-rectal-cancer-patients-life-altering-side-effects. Accessed 21 June 2022
Rogers, A., Gibbons, D., Hanly, A., et al.: Prognostic significance of tumor budding in rectal cancer biopsies before neoadjuvant therapy. Mod. Pathol. 27, 156–162 (2014). https://doi.org/10.1038/modpathol.2013.124
Shao, Z., et al.: Transmil: transformer based correlated multiple. instance learning for whole slide image classication. CoRR (2021). arxiv.org/abs/2106.00908
Sharma, Y., Shrivastava, A., Ehsan, L., Moskaluk, C.A., Syed, S., Brown, D.E.: Cluster-to-conquer: a framework for end-to-end multi-instance learning for whole slide image classification (2021). https://doi.org/10.48550/arxiv.2103.10626
Sirinukunwattana, K., et al.: Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut 70(3), 544–554 (2021). https://doi.org/10.1136/gutjnl-2019-319866,https://gut.bmj.com/content/70/3/544
Zhang, F., Yao, S., Li, Z., et al.: Predicting treatment response to neoadjuvant chemoradiotherapy in local advanced rectal cancer by biopsy digital pathology image features. Clin. Transl. Med. 10(2), e110 (2020). https://doi.org/10.1002/ctm2.110
Acknowledgements
Financial support: RW, AS - EPSRC Center for Doctoral Training in Health Data Science (EP/S02428X/1); RW - Oxford CRUK Cancer Centre; VHK - Promedica Foundation (F-87701-41-01) and Swiss National Science Foundation (P2SKP3\(\_\)168322/1, P2SKP3\(\_\)168322/2); TSM - The Stratification in Colorectal Cancer Consortium (S:CORT) funded by the Medical Research Council and Cancer Research UK (MR/M016587/1); JR, KS - Oxford NIHR National Oxford Biomedical Research Centre and the PathLAKE consortium (InnovateUK). The ARISTOTLE trial was funded by Cancer Research UK (CRUK/08/032). The computational aspects of this research were funded from the NIHR Oxford BRC with additional support from the Wellcome Trust Core Award Grant Number 203141/Z/16/Z. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Wood, R. et al. (2022). Enhancing Local Context of Histology Features in Vision Transformers. In: Kakileti, S.T., et al. Artificial Intelligence over Infrared Images for Medical Applications and Medical Image Assisted Biomarker Discovery. MIABID AIIIMA 2022 2022. Lecture Notes in Computer Science, vol 13602. Springer, Cham. https://doi.org/10.1007/978-3-031-19660-7_15
Download citation
DOI: https://doi.org/10.1007/978-3-031-19660-7_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-19659-1
Online ISBN: 978-3-031-19660-7
eBook Packages: Computer ScienceComputer Science (R0)