Abstract
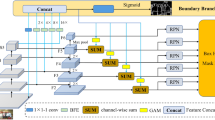
Top-down instance segmentation framework has shown its superiority in object detection compared to the bottom-up framework. While it is efficient in addressing over-segmentation, top-down instance segmentation suffers from over-crop problem. However, a complete segmentation mask is crucial for biological image analysis as it delivers important morphological properties such as shapes and volumes. In this paper, we propose a region proposal rectification (RPR) module to address this challenging incomplete segmentation problem. In particular, we offer a progressive ROIAlign module to introduce neighbor information into a series of ROIs gradually. The ROI features are fed into an attentive feed-forward network (FFN) for proposal box regression. With additional neighbor information, the proposed RPR module shows significant improvement in correction of region proposal locations and thereby exhibits favorable instance segmentation performances on three biological image datasets compared to state-of-the-art baseline methods. Experimental results demonstrate that the proposed RPR module is effective in both anchor-based and anchor-free top-down instance segmentation approaches, suggesting the proposed method can be applied to general top-down instance segmentation of biological images. Code is available (https://github.com/qzhangli/RPR).
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Chang, Q., et al.: Deeprecon: Joint 2d cardiac segmentation and 3d volume reconstruction via a structure-specific generative method. arXiv preprint arXiv:2206.07163 (2022)
Chen, H., Qi, X., Yu, L., Dou, Q., Qin, J., Heng, P.A.: DCAN: deep contour-aware networks for object instance segmentation from histology images. Med. Image Anal. 36, 135–146 (2017)
Gao, Y., Zhou, M., Liu, D., Yan, Z., Zhang, S., Metaxas, D.: A data-scalable transformer for medical image segmentation: architecture, model efficiency, and benchmark. arXiv preprint arXiv:2203.00131 (2022)
Gao, Y., Zhou, M., Metaxas, D.N.: UTNet: a hybrid transformer architecture for medical image segmentation. In: de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C. (eds.) MICCAI 2021. LNCS, vol. 12903, pp. 61–71. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87199-4_6
Girshick, R.: Fast R-CNN. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1440–1448 (2015)
He, K., Gkioxari, G., Dollár, P., Girshick, R.: Mask R-CNN. In: Proceedings of the IEEE International Conference on Ccomputer Vision, pp. 2961–2969 (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
He, X., Tan, C., Qiao, Y., Tan, V., Metaxas, D., Li, K.: Effective 3d humerus and scapula extraction using low-contrast and high-shape-variability MR data. In: Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging. vol. 10953, p. 109530O. International Society for Optics and Photonics (2019)
Hu, J.B., Guan, A., Zhangli, Q., Sayadi, L.R., Hamdan, U.S., Vyas, R.M.: Harnessing machine-learning to personalize cleft lip markings. Plastic Reconstruct. Surgery-Global Open 8(9S), 150–151 (2020)
Irshad, H., et al.: Crowdsourcing image annotation for nucleus detection and segmentation in computational pathology: evaluating experts, automated methods, and the crowd. In: Pacific Symposium on Biocomputing Co-chairs, pp. 294–305. World Scientific (2014)
Lee, Y., Park, J.: Centermask: real-time anchor-free instance segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 13906–13915 (2020)
Lin, T.Y., Dollár, P., Girshick, R., He, K., Hariharan, B., Belongie, S.: Feature pyramid networks for object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2117–2125 (2017)
Liu, D., Gao, Y., Zhangli, Q., Yan, Z., Zhou, M., Metaxas, D.: Transfusion: multi-view divergent fusion for medical image segmentation with transformers. arXiv preprint arXiv:2203.10726 (2022)
Liu, D., Ge, C., Xin, Y., Li, Q., Tao, R.: Dispersion correction for optical coherence tomography by the stepped detection algorithm in the fractional Fourier domain. Opt. Express 28(5), 5919–5935 (2020)
Liu, D., Liu, J., Liu, Y., Tao, R., Prince, J.L., Carass, A.: Label super resolution for 3d magnetic resonance images using deformable u-net. In: Medical Imaging 2021: Image Processing, vol. 11596, p. 1159628. International Society for Optics and Photonics (2021)
Liu, D., Xin, Y., Li, Q., Tao, R.: Dispersion correction for optical coherence tomography by parameter estimation in fractional Fourier domain. In: 2019 IEEE International Conference on Mechatronics and Automation (ICMA), pp. 674–678. IEEE (2019)
Liu, D., Yan, Z., Chang, Q., Axel, L., Metaxas, D.N.: Refined deep layer aggregation for multi-disease, multi-view & multi-center cardiac MR segmentation. In: Puyol Antón, E., et al. (eds.) STACOM 2021. LNCS, vol. 13131, pp. 315–322. Springer, Cham (2022). https://doi.org/10.1007/978-3-030-93722-5_34
Minervini, M., Fischbach, A., Scharr, H., Tsaftaris, S.A.: Finely-grained annotated datasets for image-based plant phenotyping. Pattern Recogn. Lett. 81, 80–89 (2016)
Oda, H., et al.: BESNet: boundary-enhanced segmentation of cells in histopathological images. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 228–236. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_26
Pantanowitz, L.: Digital images and the future of digital pathology. J. Pathol. Inform. 1, 15 (2010)
Payer, C., Štern, D., Neff, T., Bischof, H., Urschler, M.: Instance segmentation and tracking with cosine embeddings and recurrent hourglass networks. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 3–11. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_1
Ren, S., He, K., Girshick, R., Sun, J.: Faster R-CNN: towards real-time object detection with region proposal networks. In: Advances in Neural Information Processing Systems, vol. 28 (2015)
Schmidt, U., Weigert, M., Broaddus, C., Myers, G.: Cell detection with star-convex polygons. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 265–273. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_30
Tian, Z., Shen, C., Chen, H., He, T.: FCOS: fully convolutional one-stage object detection. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 9627–9636 (2019)
Vaswani, A., et al.: Attention is all you need. In: Advances in Neural Information Processing Systems, vol. 30 (2017)
Yi, J., et al.: Object-guided instance segmentation for biological images. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 34, pp. 12677–12684 (2020)
Yi, J., et al.: Multi-scale cell instance segmentation with keypoint graph based bounding boxes. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11764, pp. 369–377. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32239-7_41
Yi, J., Wu, P., Jiang, M., Huang, Q., Hoeppner, D.J., Metaxas, D.N.: Attentive neural cell instance segmentation. Med. Image Anal. 55, 228–240 (2019)
Zhang, Y., et al.: A multi-branch hybrid transformer network for corneal endothelial cell segmentation. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12901, pp. 99–108. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87193-2_10
Zhou, Y., Onder, O.F., Dou, Q., Tsougenis, E., Chen, H., Heng, P.-A.: CIA-Net: robust nuclei instance segmentation with contour-aware information aggregation. In: Chung, A.C.S., Gee, J.C., Yushkevich, P.A., Bao, S. (eds.) IPMI 2019. LNCS, vol. 11492, pp. 682–693. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-20351-1_53
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Zhangli, Q. et al. (2022). Region Proposal Rectification Towards Robust Instance Segmentation of Biological Images. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. MICCAI 2022. Lecture Notes in Computer Science, vol 13434. Springer, Cham. https://doi.org/10.1007/978-3-031-16440-8_13
Download citation
DOI: https://doi.org/10.1007/978-3-031-16440-8_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16439-2
Online ISBN: 978-3-031-16440-8
eBook Packages: Computer ScienceComputer Science (R0)