Abstract
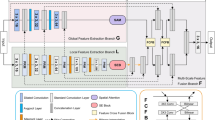
Medical image segmentation is one of the most fundamental tasks concerning medical information analysis. Various solutions have been proposed so far, including many deep learning-based techniques, such as U-Net, FC-DenseNet, etc. However, high-precision medical image segmentation remains a highly challenging task due to the existence of inherent magnification and distortion in medical images as well as the presence of lesions with similar density to normal tissues. In this paper, we propose TFCNs (Transformers for Fully Convolutional denseNets) to tackle the problem by introducing ResLinear-Transformer (RL-Transformer) and Convolutional Linear Attention Block (CLAB) to FC-DenseNet. TFCNs is not only able to utilize more latent information from the CT images for feature extraction, but also can capture and disseminate semantic features and filter non-semantic features more effectively through the CLAB module. Our experimental results show that TFCNs can achieve state-of-the-art performance with dice scores of 83.72% on the Synapse dataset. In addition, we evaluate the robustness of TFCNs for lesion area effects on the COVID-19 public datasets. The Python code will be made publicly available on https://github.com/HUANGLIZI/TFCNs.
Z. Li and D. Li—Means equal contribution.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Carion, N., Massa, F., Synnaeve, G., Usunier, N., Kirillov, A., Zagoruyko, S.: End-to-end object detection with transformers. In: Vedaldi, A., Bischof, H., Brox, T., Frahm, J.-M. (eds.) ECCV 2020. LNCS, vol. 12346, pp. 213–229. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-58452-8_13
Chen, J., et al.: Transunet: transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021)
Chowdhury, G.G.: Natural language processing. Ann. Rev. Inf. Sci. Technol. 37(1), 51–89 (2003)
Dosovitskiy, A., et al.: An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020)
He, K., Zhang, X., Ren, S., Sun, J.: Identity mappings in deep residual networks. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9908, pp. 630–645. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46493-0_38
Hendrycks, D., Gimpel, K.: Gaussian error linear units (GELUs). arXiv preprint arXiv:1606.08415 (2016)
Iandola, F., Moskewicz, M., Karayev, S., Girshick, R., Darrell, T., Keutzer, K.: Densenet: implementing efficient convnet descriptor pyramids. arXiv preprint arXiv:1404.1869 (2014)
Jégou, S., Drozdzal, M., Vazquez, D., Romero, A., Bengio, Y.: The one hundred layers tiramisu: fully convolutional densenets for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, pp. 11–19 (2017)
LeCun, Y., Bottou, L., Bengio, Y., Haffner, P.: Gradient-based learning applied to document recognition. Proc. IEEE 86(11), 2278–2324 (1998)
Li, Z., et al.: LViT: language meets vision transformer in medical image segmentation. arXiv preprint arXiv:2206.14718 (2022)
Litjens, G., et al.: Evaluation of prostate segmentation algorithms for MRI: the promise12 challenge. Med. Image Anal. 18(2), 359–373 (2014)
Liu, Z., et al.: Swin transformer: hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 10012–10022 (2021)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Oktay, O., et al.: Attention u-net: learning where to look for the pancreas. arXiv preprint arXiv:1804.03999 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Tomar, N.K., et al.: DDANet: dual decoder attention network for automatic polyp segmentation. In: Del Bimbo, A., et al. (eds.) ICPR 2021. LNCS, vol. 12668, pp. 307–314. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-68793-9_23
Valanarasu, J.M.J., et al.: Medical transformer: gated axial-attention for medical image segmentation. arXiv preprint arXiv:2102.10662 (2021)
Vaswani, A., et al.: Attention is all you need. In: Advances in Neural Information Processing Systems, vol. 30 (2017)
Wang, C.S., Su, F.Y., Lee, T.L.M., Tsai, Y.S., Chiang, J.H.: CUAB: convolutional uncertainty attention block enhanced the chest x-ray image analysis. arXiv preprint arXiv:2105.01840 (2021)
Wang, G., Li, W., Aertsen, M., Deprest, J., Ourselin, S., Vercauteren, T.: Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing 338, 34–45 (2019)
Woo, S., Park, J., Lee, J.Y., Kweon, I.S.: CBAM: convolutional block attention module. In: Proceedings of the European Conference on Computer Vision (ECCV), pp. 3–19 (2018)
Xiao, X., Lian, S., Luo, Z., Li, S.: Weighted res-unet for high-quality retina vessel segmentation. In: 2018 9th International Conference on Information Technology in Medicine and Education (ITME), pp. 327–331. IEEE (2018)
Zhang, R., et al.: Automatic segmentation of acute ischemic stroke from DWI using 3-D fully convolutional densenets. IEEE Trans. Med. Imaging 37(9), 2149–2160 (2018)
Zheng, S., et al.: Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 6881–6890 (2021)
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., Torralba, A.: Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2921–2929 (2016)
Zhou, D., et al.: Deepvit: towards deeper vision transformer. arXiv preprint arXiv:2103.11886 (2021)
Zhou, Z., Rahman Siddiquee, M.M., Tajbakhsh, N., Liang, J.: UNet++: a nested U-Net architecture for medical image segmentation. In: Stoyanov, D., et al. (eds.) DLMIA/ML-CDS -2018. LNCS, vol. 11045, pp. 3–11. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00889-5_1
Acknowledgement
This work was supported in part by the Natural Science Foundation of Fujian Province of China (No. 2020J01006), the National Natural Science Foundation of China (No. 61502402), and the Open Project Program of State Key Laboratory of Virtual Reality Technology and Systems, Beihang University (No. VRLAB2022AC04).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Li, Z. et al. (2022). TFCNs: A CNN-Transformer Hybrid Network for Medical Image Segmentation. In: Pimenidis, E., Angelov, P., Jayne, C., Papaleonidas, A., Aydin, M. (eds) Artificial Neural Networks and Machine Learning – ICANN 2022. ICANN 2022. Lecture Notes in Computer Science, vol 13532. Springer, Cham. https://doi.org/10.1007/978-3-031-15937-4_65
Download citation
DOI: https://doi.org/10.1007/978-3-031-15937-4_65
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-15936-7
Online ISBN: 978-3-031-15937-4
eBook Packages: Computer ScienceComputer Science (R0)