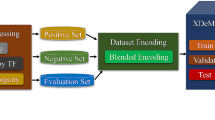
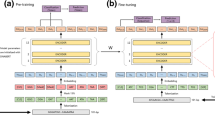
Abstract
Deciphering the language of DNA has always been one of the difficult problems that informatics methods need to deal with. In order to meet this challenge, many deep learning models have been proposed. Among them, DNA-language models based on pre-trained Bidirectional Encoder Representations from Transformers (BERT) is one of the methods with excellent performance in recognition accuracy. At the same time, most studies focus on the design of the model structure, while for pre-trained DNA-language models such as BERT, there are relatively few studies on the influence of the fine-tuning stage on model performance. To this end, we select DNABERT, the first pre-trained BERT model for DNA-language, to analysis its fine-tuning performances with different parameters settings in motif mining tasks, which are one of the most classic missions for prediction of DNA sequence binding specificities. Furthermore, we compare the fine-tuning results to the performances of previously existing models by dividing different types of datasets. The results show that in fine-tuning phase, different hyper-parameters combinations and types of dataset do have significant impact on model performance.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
D'haeseleer, P.: What are DNA sequence motifs? Nat. Biotechnol. 24, 423–425 (2006)
Nirenberg, M., Leder, P.: RNA codewords and protein synthesis, VII. On the general nature of the RNA code. Proc. Natl. Acad. Sci. USA 53, 1161–1168 (1965)
Galas, D.J., Schmitz, A.: DNAase footprinting a simple method for the detection of protein-DNA binding specificity. Nucleic. Acids Res. 5(9), 3157–3170 (1978)
Hellman, L., Fried, M.: Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat. Protoc. 2, 1849–1861 (2007)
Schenborn, E., Groskreutz, D.: Reporter gene vectors and assays. Mol. Biotechnol. 13, 29–44 (1999)
Trabelsi, A., Chaabane, M., Ben-Hur, A.: Comprehensive evaluation of deep learning architectures for prediction of DNA/RNA sequence binding specificities. Bioinformatics 35(14), i269–i277 (2019)
LeCun, Y.: Gradient-based learning applied to document recognition. Proc. IEEE. 86, 2278–2324 (1998)
Hochreiter, S., Schmidhuber, J.: Long short-term memory. Neural Comput. 9, 1735–1780 (1997)
Alipanahi, B., Delong, A., Weirauch, M.: Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 (2015)
Zhu, L., Zhang, H.B., Huang, D.S.: Direct AUC optimization of regulatory motifs. Bioinformatics 33(14), i243–i251 (2017)
Shen, Z., Zhang, Y.H., Han, K.S., Nandi, A.K., Honig, B., Huang, D.S.: miRNA-disease association prediction with collaborative matrix factorization. Complexity. 2017(2017), 1–9 (2017)
Gupta, A., Rush, A.M.: Dilated convolutions for modeling long-distance genomic dependencies. arXiv:1710.01278 (2017)
Davuluri, R.V.: The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 24, 167–177 (2008)
Gibcus, J.H., Dekker, J.: The context of gene expression regulation. F1000 Biol. Rep. 4, 8 (2012)
Vitting-Seerup, K., Sandelin, A.: The landscape of isoform switches in human cancers. Mol. Cancer Res. 15, 1206–1220 (2017)
Zhang, H.B., Zhu, L., Huang, D.S.: WSMD: weakly-supervised motif discovery in transcription factor ChIP-seq data. Sci. Rep. 7 (2017)
Quang, D., Xie, X.: A hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 44, e107 (2016)
Zhou, Y.X., Hefenbrock, M., Huang, Y.R., Riedel, T., Beigl, M.: Automatic Remaining Useful Life Estimation Framework with Embedded Convolutional LSTM as the Backbone. ECML PKDD 2020: Machine Learning and Knowledge Discovery in Databases: Applied Data Science Track, pp. 461–477 (2020)
Schuster, M., Paliwal, K.K.: Bidirectional recurrent neural networks. IEEE Trans. Sig. Process. 45(11), 2673–2681 (1997)
Vaswani, A., et al.: Attention is all you need. In: Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS 2017) (2017)
Devlin, J., Chang, M.W., Lee, K., Toutanova, K.: BERT: pre-training of Deep Bidirectional Transformers for Language Understanding (2018)
Taylor, W.L.: Cloze procedure: a new tool for measuring readability. J. Bull. 30(4), 415–433 (1953)
Wang, A., Singh, A., Michael, J., Hill, F., Levy, O., Bowman, S.: Glue: a multi-task benchmark and analysis platform for natural language understanding. In: Proceedings of the 2018 EMNLP Workshop BlackboxNLP: Analyzing and Interpreting Neural Networks for NLP. 2018a, pp. 353–355 (2018)
Rajpurkar, P., Zhang, J., Lopyrev, K., Liang, P.: Squad: 100,000+ questions for machine comprehension of text. In: Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, pp. 2383–2392(2016)
Zhu, L., Zhang, H.B., Huang, D.S.: LMMO: a large margin approach for optimizing regulatory motifs. IEEE/ACM Trans. Comput. Biol. Bioinf. 15(3), 913–925 (2018)
Ji, Y.R., Zhou, Z.H., Liu, H., Davuluri, R.V.: DNABERT: pre-trained bidirectional encoder representations from transformers model for DNA-language in genome. Bioinformatics 37(15), 2112–2120 (2021)
Dreos, R., Ambrosini, G., Périer, R.C., Bucher, P.: EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res. 41(D1), D157–D164 (2013)
Oubounyt, M., Louadi, Z., Tayara, H., Chong, K.T.: DeePromoter: robust promoter predictor using deep learning. Front Genet. 10, 286 (2019)
Zhang, H.B., Zhu, L., Huang, D.S.: DiscMLA: An efficient discriminative motif learning algorithm over high-throughput datasets. IEEE/ACM Trans. Comput. Biol. Bioinf. 15(6), 1810–1820 (2018)
Solovyev, V., Kosarev, P., Seledsov, I.: Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7(S10) (2006)
Davuluri, R.V.: Application of FirstEF to find promoters and first exons in the human genome. Current Protocols Bioinform. 1, 4.7.1–4.7.10 (2003)
The ENCODE Project Consortium: An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012)
Zhang, Y., Qiao, S., Ji, S., Li, Y.: DeepSite: bidirectional LSTM and CNN models for predicting DNA–protein binding. Int. J. Mach. Learn. Cybern. 11(4), 841–851 (2019). https://doi.org/10.1007/s13042-019-00990-x
Khamis, A.M., et al.: A novel method for improved accuracy of transcription factor binding site prediction. Nucleic Acids Res. 46(12), e72 (2018)
Shen, Z., Zhang, Q., Han, K., Huang, D.S.: A deep learning model for RNA-protein binding preference prediction based on hierarchical LSTM and attention network. IEEE/ACM Trans. Comput. Biol. Bioinform. 19 (2020)
Zhang, Q., Shen, Z., Huang, D.S.: Predicting in-vitro transcription factor binding sites using DNA sequence shape. IEEE/ACM Trans. Comput. Biol. Bioinform. 18 (2019)
Shen, Z., Deng, S.P., Huang, D.S.: Capsule network for predicting RNA-Protein binding preferences using hybrid feature. IEEE/ACM Trans. Comput. Biol. Bioinform. 17 (2019)
Zhu, L., Bao, W.Z., Huang, D.S.: Learning TF binding motifs by optimizing fisher exact test score. IEEE/ACM Trans. Comput. Biol. Bioinform. (2016)
Shen, Z., Deng, S.P., Huang, D.S.: RNA-Protein binding sites prediction via multi-scale convolutional gated recurrent unit networks. IEEE Trans. Comput. Biol. Bioinform. 17 (2019)
Zhang, Q.H., Zhu, L., Bao, W.Z., Huang, D.S.: Weakly-supervised convolutional neural network architecture for predicting protein-DNA binding. IEEE/ACM Trans. Comput. Biol. Bioinform. 17 (2020)
Zhang, Q.H., Zhu, L., Huang, D.S.: High-order convolutional neural network architecture for predicting DNA-protein binding sites. IEEE/ACM Trans. Comput. Biol. Bioinform. 16 (2019)
Zhang, Q.H., Shen, Z., Huang, D.S.: Modeling in-vivo protein-DNA binding by combining multiple-instance learning with a hybrid deep neural network. Sci. Rep. 9, 8484 (2019)
Xu, W.X., Zhu, L., Huang, D.S.: DCDE: an efficient deep convolutional divergence encoding method for human promoter recognition. IEEE Trans. Nanobiosci. 18(2), 136–145 (2019)
Shen, Z., Bao, W.Z., Huang, D.S.: Recurrent neural network for predicting transcription factor binding sites. Sci. Rep. 8, 15270 (2018)
Acknowledgements
This work was supported by the grant of National Key R&D Program of China (No. 2018YFA0902600 & 2018AAA0100100) and partly supported by National Natural Science Foundation of China (Grant nos. 61732012, 62002266, 61932008, and 62073231), and Introduction Plan of High-end Foreign Experts (Grant no. G2021033002L) and, respectively, supported by the Key Project of Science and Technology of Guangxi (Grant no. 2021AB20147), Guangxi Natural Science Foundation (Grant nos. 2021JJA170204 & 2021JJA170199) and Guangxi Science and Technology Base and Talents Special Project (Grant nos. 2021AC19354 & 2021AC19394).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Tan, X., Yuan, C., Wu, H., Zhao, X. (2022). Comprehensive Evaluation of BERT Model for DNA-Language for Prediction of DNA Sequence Binding Specificities in Fine-Tuning Phase. In: Huang, DS., Jo, KH., Jing, J., Premaratne, P., Bevilacqua, V., Hussain, A. (eds) Intelligent Computing Theories and Application. ICIC 2022. Lecture Notes in Computer Science, vol 13394. Springer, Cham. https://doi.org/10.1007/978-3-031-13829-4_8
Download citation
DOI: https://doi.org/10.1007/978-3-031-13829-4_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-13828-7
Online ISBN: 978-3-031-13829-4
eBook Packages: Computer ScienceComputer Science (R0)