Abstract
Plant genetic resources are source genetic material for conducting research and breeding. The use of this material is subject to international and national regulations on access and benefit-sharing (ABS). With modern genetic technologies generating desired trait and gene function improvement by replicating genetic signatures, ABS must adapt to the new technological reality. As the constituencies of international ABS conventions discuss if and how to extend the application of the conventions to digital sequence information (DSI) derived from source material, the genomics science community resists any incumbrance to continued free and unrestricted access to such information. Based on current ABS discussions and the likely future co-existence of diverse policy regimes, this paper proposes interoperability among data systems as an essential tool to implement legal solutions for benefit-sharing as well as advance science and innovation objectives. Two information technology tools are suggested for associating DSI to plant genetic resources and reciprocal citations with data exchange, namely digital object identifiers and digital genetic objects. This paper concludes that interoperability should be experimented with in both its technical and social dimensions, in order to support long-term alliances between policy and science through data archives, knowledge bases and live specimen collection resources.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
1 Introduction
In plant biology research and crop breeding programs, the value of plant genetic resources is determined by the seed and the propagation material, called source genetic material, that are important for conducting genomics, genetics, phenotype and trait evaluation, in-vivo and in-vitro experiments. Much of the experimental information, data, and the knowledge gained become important for the researchers when they are properly associated with the source genetic material, thus enabling further scientific discovery and future replication of the studies. Often, the physical plant material and the derivatives (including isolated protein, DNA and RNA) are however difficult to access due to various national and international regulations and exchange permits. On the one hand, this limited access restricts the use of existing genetic material; whereas on the other hand it can require more tracking of use and citing the source material for various purposes, including publication. Associating experimental information with its source genetic material begins to bring aspects of plant science within the purview of global agreements that establish rules for accessing the source genetic material for research and development and sharing the benefits of its utilization. Under one such agreement, namely the International Treaty on Plant Genetic Resources for Food and Agriculture (ITPGRFA), a Global Information System (GLIS) was established to facilitate the exchange of information on crop genetic material.Footnote 1
Innovation at the intersection of digital technologies and life sciences is quickly changing the context of the global agreements on genetic resources. Modern bioscience relies on the extraction and processing of large volumes of “omics’’ data in digital form, and this has precipitated a re-examination of the founding principles of such global agreements, as they relate to matters such as identification of the resource, monitoring of its use and attribution of the benefits of such use (Aubry, 2019; Welch et al., 2017). While whole-genome sequences are increasingly available as a result of new-generation technologies, the collective capacity to actually analyze and benefit from the data is lagging behind (Halewood et al., 2018).
The interaction between policymakers driving the global agreements on genetic resources and the genomics science community can be problematic. In the governance frameworks of the global agreements, this interaction is viewed through the lens of digital sequence information (DSI), a term of uncertain meaning that functions as a placeholder in the discussions as to whether the informational component of genetic resources should be regulated under the same rules of access and benefit-sharing (ABS) that govern source genetic material.
The respective value propositions seem to radically differ. ABS is equity-driven and relies on normative standards (legislation, contracts) to implement controlled access regimes (Ruiz, 2015). The genomics science community prioritizes research efficiency and is guided by community standards and protocols, e.g. the Fort Lauderdale agreement, the Toronto agreement, FAIR data principles on data sharing with international archives and publications (Toronto International Data Release Workshop Authors, 2003; Wellcome Trust, 2003; Wilkinson et al., 2016). According to these standards, all genomics data including derived DNA, RNA and protein sequences must remain public and accessible without restrictions in order to enable biologists to discover and realize the benefits of the material in research and application.Footnote 2 The access to derived DNA, RNA and protein sequences from the physical genetic material has opened up a new possibility in the research and innovation community enabled by genetic engineering technologies like CRISPR (Chen et al., 2019). Now, researchers have the ability to update genomes of a germplasm by replicating genetic signatures of a wild relative with sequences associated with desired trait and/or gene function improvements, without actually accessing the original seed material considered a global and national heritage. Thus, new proposals and insights are under discussion to revisit the mandates of ABS international agreements for protecting the community interests that take into account the related compensatory gains derived from the genetic signatures and sequences to achieve genetic gains and trait improvements.
In our paper, we introduce ABS policy discussions around DSI, and argue that interoperability among data systems will be essential to implement future legal solutions for benefit-sharing. With a view to pursuing such interoperability, we suggest possible mechanisms that may be well-aligned with the spirit of the international agreements, to develop optimal and timely recommendations for associating DSI to the plant genetic resources and reciprocal citations with data exchange. One mechanism is based on the integration between the federated system of databases of the International Nucleotide Sequence Database Collaboration (INSDC) and the current tools that are available to the plant science community through the GLIS. Another mechanism revolves around the proposed concept of Digital Genetic Object (DGO) as a way to introduce a precise definition of DSI that is functional to interoperability among biological data systems. In conclusion, we flag the need to continue approaching data interoperability with a dual focus on global policy and information technology.
2 Global Policy on Access and Benefit-Sharing and the Nexus with Interoperability
ABS is a construct of international agreements on genetic resources. In Article 2 of the Convention on Biological Diversity, genetic resources are defined as any material of plant, animal, microbial or other origin containing functional units of heredity, of actual or potential value.Footnote 3 ABS is aimed at exploiting, through controlled access, the potential of those resources for various public policy objectives, e.g. nature conservation, food security, sustainable development, and at rewarding, through the fair and equitable sharing of the benefits of utilization, those who maintain the diverse genetic base.
In various ABS international fora, including the ITPGRFA at the Food and Agriculture Organization of the United Nations, discussions are taking place as to whether to regulate DSI within the remit of the agreements. The motivation to subsume DSI into the domain of ABS, is to realize the provisions of the agreements in the light of scientific and technological advancements. Thanks to such advancements, innovation increasingly relies on the intangible component of genetic resources, i.e. information and data. Although at present, the use of both tangible and intangible components of genetic resources co-exists, it is postulated that in the near future, additional detachment of the informational component from the physical organisms will occur (Morgera et al., 2020; Smyth et al., 2020).
The priority focus of the ABS community is on three issues. The first is the scope of DSI, that is, the data sets that DSI encompasses. The scope of DSI is still under consideration and options range from only the base sequence of genomic DNA to all information associated with genetic resources. Being cognizant of such a broad range of options, our examination considers categories of data which may fit into a functional definition of DSI, namely: DNA, RNA, protein, genetic markers (with or without sequences), non-coding features and other data categories (Houssen et al., 2020; Brink et al., 2021) and specifically suggests ways to link these data to other ontologized knowledge to accommodate expansive views of DSI. As best practice, existing ontology may be used where each concept bears a unique and resolvable identifier, called a Uniform Resource Identifier, for which the definition, context of use and semantic relationships are validated by a large community (Arnaud et al., 2020). For example, the Sequence Ontology, the Protein Ontology, and the Gene Ontology, which include concepts and definitions of Genomic Objects along with other relevant ontologies, such as the NCBI taxonomy for species, and metadata standards, such as the Biosample record, may provide a useful point of departure.Footnote 4,Footnote 5 As discussed in another chapter of this book, cross-domain ontologies have the potential to reduce concept proliferation (see Devare et al).The second issue is terminology, that is, a scientifically accurate term that can be applied in the governance of the international legal agreements. Terms that are under consideration include genetic sequence data, genomics information, natural information (Convention on Biological Diversity, 2020). The third issue is traceability of DSI in databases and in research and development activities that utilize DSI (Convention on Biological Diversity cit.). Traceability relates data to a particular genetic resource or to any source that implies the utilization of a genetic resource.
While scientists continue to rely on open access to sequence data, ABS policy demands benefit-sharing and brings this open system into question (Rohden et al., 2021). In the course of such discussions, policy options for utilization of DSI and benefit-sharing have begun emerging. The spectrum of such options is ample. It ranges from free and unrestricted access to genomics data coupled with the financing of benefit-sharing through a multilateral fund, to controlled access to databases and a transactional approach to benefit-sharing, with “club-approach” solutions, such as membership or cloud-based fees, commons licenses, also being proposed (Hartman-Sholz et al., 2020).Footnote 6
Once the ABS policy discussions are complete, the expectation is that a functional definition of DSI will be agreed upon and solutions will be put in place in the framework of the ABS agreements to address the utilization of DSI and benefit-sharing. The current fragmentation of the global ABS framework illustrates an example of a regime complex, with overlapping institutions that interact among themselves on patterns of hierarchy and differentiation (Randall Henning et al., 2020). In the light of such institutional complexity and given the plurality of policy options that are being discussed for DSI, it is likely that different solutions will go through an initial phase of experimentation and thus co-exist, e.g. for different categories of genetic resources and derived DSI. By way of example, some genetic resources and the derived DSI may be reserved to national sovereignty and the ensuing control of access and use, and others may be grouped into one or multiple global pools and administered in accordance with open access standards, coupled with multilateral benefit-sharing mechanisms, including pursuant to Article 10 of the Nagoya Protocol.Footnote 7
As diverse policy options are likely to co-exist, it is foreseeable that data aggregation and interoperability will play a key role in implementing corresponding solutions for benefit-sharing. Identifying data sets as DSI and associating DSI to defined genetic resources will be necessary to impute individual or aggregate benefits to the use of identified data and resources.
A number of data sets that are under consideration as DSI are stored and accessed in a variety of databases including the INSDC (Rohden et al., 2021). For the international policy decisions on the scope of DSI to be channeled to actual producers and users of sequence data annotated with DSI, harmonization with the database system and the underlying technology and standards will be highly desirable, if not indispensable. In practice, sequence databases will need to be interoperable with each other in order to identify DSI for legal purposes. The digital nature of sequence information renders it mandatory to propose, in parallel to the current legal discussion, solutions for the interoperability of the data to complement decisions about ABS. Clear, precise definitions of types of genetic material and data must be put into practice through improved data aggregation and interoperability, and increased integration among information systems.
As improved data curation, standardization, identification of provenance, aggregation, exchange and interoperability may support the unfolding institutional processes related to DSI, the outcomes of such processes are likely to elicit varied responses by the science community, based on different assumptions about the degree of choice, awareness, and self-interest. Academic scientists’ responses to new regulatory controls on biological material inputs to research show a degree of variation that is shaped by both micro-level, cognitive and macro-level, institutional factors (Oliver, 1991; Welch et al., 2019). Within such a spectrum of responses, some researchers may not be inclined at all to support ABS policy processes in relation to DSI. Nevertheless, others may be willing to pursue anticipatory action with respect to DSI policy development, for instance to increase legitimacy and social qualification that are instrumental to resource mobilization. Such anticipatory action may offer other considerable benefits, such as exerting influence on implementation and co-opting technical standards.
3 Interoperability in the Global Information System of the International Treaty: Possible Applications to Exchanges of DSI
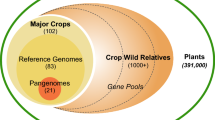
The ITPGRFA is one of the international instruments that compose the global ABS architecture. GLIS is founded on the principle of integration with existing information systems. It implements data aggregation and interoperability by associating information and knowledge to plant genetic resources in ex-situ, in-situ and on farm conditions to facilitate research and breeding for food and agriculture, as shown in Fig. 1 below. This association is pursued through permanent unique identifiers. Among the different identifier technologies considered, Digital Object Identifiers (DOIs) emerged as a very powerful mechanism to establish linkages to all sorts of information. DOIs are a well-established standard originally developed for the publication sector that has recently expanded its reach to many other application fields.Footnote 8
Among other desirable qualities, DOIs are well known in the research space, offer advanced services such as EventData and the PID Graph, are widely adopted by the publishing sector and dataset repositories, and support flexible metadata structures allowing representation of object types of very different nature. All these characteristics will come handy when we describe our proposed solution.Footnote 9
Although the ITPGRFA community has not directly tackled the complex ABS legal issues through GLIS, it has acquired experience on the technical implementation of information-sharing in the context of global policy and multilateral cooperation, and has implemented a blended approach to interoperability combining technical standards with iterative learning and multilateral dialogue (Morgera et al., 2020).Footnote 10 Such features of GLIS make it suitable to explore possible further integration options between repositories of source genetic material and large repositories of genomics data.
As mentioned above, the diversified components of DSI are likely spread across multiple repositories and databases, each one designed and refined over time to meet the demands of its own user community. The multiplicity of repositories and user requirements undoubtedly poses a variety of challenges that cannot be reduced or solved through a uniform, standard solution. The approach of this paper is to initially tackle data aggregation and interoperability through identification. The identification and ability to link the array of component parts that are all suggested as being part of DSI, depending on the definition adopted, could enable creating the relationships between those component parts and ultimately improve the information discovery and insight about the plant genetic resources themselves.
Insofar as nucleotide sequence data as well as other components of DSI can express their full value in conjunction with passport data and other information on the source genetic material, the identification of such material emerges as an actual challenge that needs solutions for the attainment of interoperability. The identification of source material is an area where many genomics repositories currently provide little precision and traceability. For example, INSDC does not offer an accurate identification of the original material, as shown in Fig. 2 below.
The “source” block under “FEATURES” at the bottom of the page is a formatted text attribute that is not mandatory. The rice cultivar name “IR64” is indeed provided but this may not be sufficient to properly identify the original material nor would be a locally assigned identifier, such as a genebank accession number, as cultivars and genebank accessions are often genetically heterogeneous.
Such deficiency may be imputed to the fact that, until the deployment of DOIs by GLIS, there has been no practical solution to accurately and permanently reference a sample of crop germplasm across information systems. During the last 3 years, DOIs have addressed the issues arising with locally assigned identifiers that may cause collisions when taken out of the assigning institution’s context. Through GLIS, assigning DOIs to plant genetic resources is a rapid process that can be performed in a variety of ways from a simple web form with a handful of mandatory attributes to powerful XML-based, system-to-system messaging.
Following the principle of requiring minimal changes to existing systems while maximizing advantages for users, one practical pathway of integration between repositories of samples of source germplasm and ISNDC genomics repositories would be to mention the original material’s DOI in the “source” feature of the Accession record establishing a proper link between DSI data sets and the original material. As genomics researchers may sometimes have to multiply source genetic material that is provided in insufficient quantity, with the risk that the genetic identity of the resulting material may be altered, a new DOI could be assigned to the material that is actually sequenced. Such a new DOI would be related to the material received, thanks to specific GLIS features, and would be cited in the Accession Number. The potential of this DOI feature is clearly not limited to the INSDC data ecosystem. It could also be deployed to establish permanent relationships among multiple data sets that the definition of DSI may comprise, and between those aggregate data sets and source genetic material.
In the INSDC scenario, when displaying the Accession detail page, the system could detect the DOI in the “source” feature and transform it, through a trivial string manipulation, into a URL to the doi.org resolver leading to the landing page associated with it. This mechanism would work irrespective of the DOI being assigned by GLIS or by any other authority and irrespective of it being associated with a plant or other lifeform. This simple transformation would already significantly improve the user experience and add real value to the Accession record.
While this minimalist approach may benefit some INSDC users, it may need complementation for other user communities that GLIS serves. In this perspective, the link to the INSDC Accession could also be provided in the GLIS DOI detail page, as shown in Fig. 3 below.
Besides providing passport information, GLIS collects links to websites where additional information on the PGRFA can be found and maintains a graph showing how the material was obtained, as illustrated in Fig. 4 below where the nodes are the DOIs associated to the materials and the arcs are the relationships linking each node to its progenitor(s). It also lists publications and datasets citing the PGRFA’s DOI. This feature is based on the EventData service, jointly developed by Crossref and DataCite,Footnote 11 and allows for automatic discovery of publications and datasets citing the current DOI.
Ideally, should INSDC opt to assign DOIs to its Accessions and properly cite the DOIs reported in the “source” feature, the link to the INSDC Accession would automatically appear in the GLIS landing page for that material thanks to Event Data. In turn, INSDC could directly benefit from Event Data services to discover publications and datasets citing the Accession’s DOI.
4 Introducing Digital Genetic Objects for Precision of Definition and Interoperability of DSI
GLIS is an enabler of a global architecture for accessing and sharing germplasm and related information. The GLIS and associated DOIs provide an approach for accurately and permanently referencing crop germplasm across information systems and, as noted above, integration of DOI into the INDSC architecture may offer tangible benefits. Currently, GLIS DOIs are assigned primarily at the genebank accession level, which can contain significant – and in many cases undiscovered – genetic diversity. An incremental option to improve interoperability relies on a finer definition and mechanisms for linking data so that GLIS DOIs can be assigned to associated DSI in a scalable and interoperable fashion. DGOs can provide this linkage.
As has been noted, there is a diversity of views of what comprises DSI. In 2018, the Ad-Hoc Technical Committee (AHTEG) of the Convention on Biological Diversity considered the following information (Convention on Biological Diversity, 2018):
-
1.
The nucleic acid sequence reads and the associated data;
-
2.
Information on the sequence assembly, its annotation and genetic mapping;
-
3.
Information on gene expression;
-
4.
Data on macromolecules and cellular metabolites;
-
5.
Information on ecological relationships and abiotic factors of the environment;
-
6.
Function, such as behavioral data;
-
7.
Structure, including morphological data and phenotype;
-
8.
Information related to taxonomy;
-
9.
Modalities of use.
In preparation for a new meeting of the AHTEG, four possible cumulative groups of information were categorized (Houssen et al. cit.):
-
1.
Narrow: DNA and RNA
-
2.
Intermediate: DNA, RNA and proteins
-
3.
Intermediate: DNA, RNA, proteins and metabolites
-
4.
Broad: DNA, RNA, protein, metabolites, germplasms, in situ and in vitro genetic material, genetic diversity, markers (genetic and molecular), microbiome, traditional knowledge, ecological interactions (Fig. 5).
In 2020, the AHTEG considered the first three groups as possibly constituting DSI and excluded the fourth group (Convention on Biological Diversity, 2020). We argue that a new identifier for DSI in information systems – DGOs – can help both the ABS and genomic science communities manage the complexity of DSI at the level of multiple data sets and association with specific PGRFA and be equipped with new approaches to facilitate implementation of policy decisions on the scope of DSI.
Modified from Houssen et al. (2020), who clustered Digital Sequence Information (DSI) into four possible cumulative groups of information
DGOs are knowledge objects created to precisely describe distinct types of DSI, objects that can be assigned GLIS DOIs and also annotated using community-driven reference vocabularies and ontologies to link to wider bodies of knowledge. Such an approach would accommodate narrow (e.g. just DNA or RNA) or broad (e.g. incorporating traditional knowledge or ecological interactions at organism, population and systems-level) definitions of DSI, and facilitate easier flow of data and knowledge across the spectrum of potential definitions.
For material for which there is an associated DOI, discrete DGOs may be created for each unique type of DSI, falling roughly within groups 1–3 of the scheme above. Each DGO can in turn then be assigned a DOI linked to the accession DOI, facilitating discovery of the associated data via GLIS and other international information systems such as INSDC. DGOs can link to a diversity of, and facilitate discovery between, bodies of knowledge related to even the broadest interpretations of what comprises DSI via data annotation leveraging reference ontologies and vocabularies. This approach points the way to describing these data in terms of their agronomic, environmental, phenotypic characteristics, and placing them more precisely in time and space to facilitate broad discovery and use of these data.
The precision of definition made possible by DGOs and the ability to link these knowledge objects to both the GLIS system through DOIs and to other bodies of knowledge through ontology-derived annotation can enable a cross-cutting ‘interoperability layer’ linking systems and existing data standards across operational domains. DGOs can represent an accelerator of scientific discovery and enhancement to public information systems through data interoperability, meeting needs of both the ABS and scientific communities.
5 Benefits and Possible Roadblocks
Despite the low investment required and the significant benefit for users of the technical options presented above, the experience with GLIS DOIs shows that there would be roadblocks to consider. First and foremost, user motivation to consistently adhere to the new workflow based on citing the original material’s DOI, including assigning a new DOI to the original material if necessary, would be increased by the immediate advantage of being able to access at least passport data available through GLIS. However, awareness will have to be raised about this new approach and its advantages.
GLIS has also experienced some unexpected setbacks when dealing with publications and datasets, which would reverberate into the INSDC association. For technical reasons, most publisher systems have difficulties in properly handling data citation, i.e. referencing DOIs not associated with bibliographic references, such as GLIS DOIs. The current solution is to list GLIS DOIs among the bibliographic references but this encounters some resistance by editors because those “references” look odd, lacking traditional elements such as title, publisher and so on. Dataset repositories, on the other hand, implement heterogeneous practices: some support data citation properly while others do not.Footnote 12
The DGO solution would have to resolve technical challenges. As the approach outlined in Fig. 6 could generate many thousands of DGOs, this will require not only precision of definition but also some operational decisions about when and how they are assigned, how to manage and store the associated data. Reference ontologies have not fully been used in this way, and would need to be fine-tuned. One initiative step will be to create a DGO ontology based on the diversity of discrete types of DGOs in groups 1–3 of DSI. Another will be to examine related reference ontologies and their suitability to linking to DGOs. In some cases, they will need to be fine-tuned to link to the material.
Digital Genetic Objects (DGO; Dark blue colored bubbles) are knowledge objects that can be assigned to distinct types of DSI, allowing more precise definition for each, their semantic relationships and derivations. DGOs can be annotated and using relevant ontologies, as well as assigned DOIs, and both methods would serve to connect DSI to wider bodies of knowledge. Currently only literature-based DOI citations hold all the unstructured information in the natural language form in the published articles. A majority carry incomplete or insufficient information and metadata to build semantic relationships between various DGOs
The pilot integration between INSDC and GLIS would pave the way for other DSI repositories towards a proper relationship between DSI and the original material. The cost/benefit ratio would be very small and would greatly improve science and the life of users of both systems. Arguably, it would also motivate users to properly reference the original material. To date, the “source” block is not much populated, likely because there is little added value in referencing the original material in a non-actionable, potentially inaccurate way.
Once interoperability with INSDC is achieved, it would be a potent success story for future extensions to any other type of database or repository of information associated to plant genetic resources registered in GLIS, leading to a coordinated constellation of systems on, for example, phenomics, traditional knowledge and technologies.
Data interoperability resulting from successful application of DGOs could provide new linkages of DSI to information systems for genetic materials, and form the basis of interoperability across research and operational domains to help build more integrated research insights and analytic infrastructures for accelerating discovery and use. Some potential high-value use-cases supported by increased data interoperability include:
-
accelerating understanding of genetic diversity within genebanks, through a cross-cutting data standard for describing results from diversity studies;
-
increased accretion of knowledge related to the material from other domains such as breeding or on-farm research;
-
revealing duplication in collections and informing the “right” level of duplication in light of long-term commitments for preservation of the genetic material;
-
precision of definition supporting data integration, in turn helping to bridge research and operational domains;
-
eased ability to compare data from multiple sequences, a key way to enhance their value (Laird & Wynberg, 2018);
-
easier linkage of data on genetic discovery generated with newer forms of measurement (e.g. multispectral imagery) and linkages of associated databases;
-
linking DSI to wider bodies of ontologized knowledge;
-
improved access to data on the complex interactions between genomics, environment, and management practices—critical for predictive modeling.
The solutions discussed in this paper in relation to plant genetic resources could apply to other biological domains (e.g. microbes, fungi, land and aquatic animals and other eukaryotes) in the INSDC collection and beyond. Establishing the connection between INSDC Accessions and the corresponding biological materials as well as applying DGOs to link DSI across taxonomic groups could be of increasing importance for synthetic biology (Rohden et al. cit.) and facilitate study of horizontal gene transfer.
In addition to these potential benefits supporting use of DSI and materials by the scientific community, DGOs appear to provide key capabilities in support of issues for the ABS community noted earlier: greater precision of definition can help with fine-tuning the terminology associated with DSI. The ability to annotate DSI (via DGOs) with diverse bodies of knowledge makes it more possible to accommodate very narrow or broad views on the scope of DSI. The ability to link precise defined and well-described data is a necessary precondition for improving overall traceability of data and the associated materials. Data standards, however, are only as good as their use in information systems, by stakeholder communities, and complex institutional contexts. Concrete pilots will be needed to test the viability of DGOs at the intersection of these dimensions.
6 Conclusion: A Common Pathway Between Global Policy on Genetic Resources and Information Technology and Data Science
The consideration of DSI by ABS policymakers requires a harmonious relationship with the genomics science community. We postulate that data aggregation and interoperability are fields where the much-needed reciprocal adjustment in processes and the blending of different rules may occur (Leonelli, 2019). Given the value of associating DSI with source genetic material, interoperability solutions should be tested based on existing genetic resource information systems. In this paper, we have suggested interoperability solutions between GLIS and INSDC as well as the introduction of DGOs into biological data systems. The insertion of DOIs into the “source” feature of the INSDC Accession record would enable relationships with passport data and other information on plant genetic resources. DGOs would further improve interoperability through a finer definition of DSI component parts and mechanisms for linking data across research and operational domains. In conjunction with these technical features, the interoperability solutions proposed in this paper would enable the smooth association of genetic resources and data in multiple repositories with applicable legal regimes governing their use.
As far as the international genomics science community that routinely manages the genetic material and the data is open to learn, develop and adopt best practices, and the genetic resources policy community seeks dialogue and cooperation, the opportunity to test and refine the two suggestions made may exist.
At the practical level, if the proposals of this paper are broadly acceptable to the scientific community, engagement with the communities maintaining relevant ontologies, metadata standards for genetic and genomic data, and annotation tools would be advisable in order to study a functional definition of DSI out of the existing ontologies, and identify gaps in both metadata and semantics in order to support interoperability of the annotated data as well as facilitate the alignment with multiple ABS policy options. Governance and oversight of this experimental system would require careful consideration in order to pursue implementation of interoperability not only as syntactic or semantic levels through data formats and communication protocols, but also as cross-domain, so to include social, policy and organizational aspects that impact on the performance of the information technology systems. This proposition resonates with the emphasis made in other chapters of this book on the key role of governance in structuring transdisciplinary collaborations across academic and non-academic communities (Louafi et al. this volume; Devare et al. this volume).
The proposals of this paper may just be one small step towards building new global standards for access and exchange of plant genetic resources and plant sequence data. Mindful of both technical opportunities and governance challenges, our hope is that this paper will be conducive to experimenting interoperability in both its technical and social dimensions, and thus represent a factual contribution in the direction of long-term alliances between policy and science through data archives, knowledge bases and live specimen collection resources.
Notes
- 1.
http://www.fao.org/3/a-i0510e.pdf, see Article 17.
- 2.
A great example is the open data sharing on COVID-19 viral genome sequences that is instrumental to developing vaccines.
- 3.
https://www.cbd.int/convention/text/, see Article 2.
- 4.
- 5.
https://fairsharing.org/biodbcore-000008/.BioSamples records include mandatory fields linked to data standards for genomic data and additional fields for particular standards.
- 6.
In this paper, the authors do not express any preference for any of the options.
- 7.
Article 10 of the Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization (ABS) to the Convention on Biological Diversity provides that “Parties shall consider the need for and modalities of a global multilateral benefit-sharing mechanism to address the fair and equitable sharing of benefits derived from the utilization of genetic resources and traditional knowledge associated with genetic resources that occur in transboundary situations or for which it is not possible to grant or obtain prior informed consent. The benefits shared by users of genetic resources and traditional knowledge associated with genetic resources through this mechanism shall be used to support the conservation of biological diversity and the sustainable use of its components globally.” https://www.cbd.int/abs/text/
- 8.
- 9.
EventData is a joint initiative of Crossref and Datacite, the two leading DOI Registration Agencies (see https://www.crossref.org/services/event-data). PID Graph is a tool funded under the EU project FREYA that collects and makes available references of DOIs to other DOIs and other PIDs (such as ORCID or ROR). See https://www.project-freya.eu/en/pid-graph/the-pid-graph
- 10.
By “ITPGRFA community”, we mean: State party delegates and the broad set of non-State actors who regularly participate in official meetings, including representatives of international and academic agricultural research, private sector, civil society, farmers.
- 11.
- 12.
References
Arnaud, E., Laporte, M. A., Kim, S., Aubert, C., Leonelli, S., Cooper, L., Jaiswal, P., Kruseman, G., Shrestha, R., Buttigieg, P. L., Mungall, C., Pietragalla, J., Agbona, A., Muliro, J., Detras, J., Hualla, V., Rathore, A., Das, R., Dieng, I., & King, B. (2020). The ontologies community of practice: An initiative by the CGIAR platform for big data in agriculture. Patterns, 1, 100–105.
Aubry, S. (2019). The future of digital sequence information for plant genetic resources for food and agriculture. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2019.01046
Convention on Biological Diversity. (2020). Report of the Ad Hoc technical expert group on digital sequence information on genetic resources. https://www.cbd.int/doc/c/911e/cc8b/de7d7fba3a8374ba4a2fbf53/dsi-ahteg-2020-01-07-en.docx. Accessed 28 Sep 2021.
Brink, M., & van Hintum, T. (2021). Practical consequences of digital sequence information (DSI) definitions and access and benefit-sharing scenarios from a plant genebank’s perspective. Plants, People, Planet. https://doi.org/10.1002/ppp3.10201
Chen, K., Wang, Y., Zhang, R., Zhang, H., & Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annual Review of Plant Biology, 70(1), 667–697.
Convention on Biological Diversity. (2018) Report of the Ad Hoc technical expert group on digital sequence information on genetic resources. https://www.cbd.int/doc/c/7ea1/36b3/7ccf849897a4c7abe49502b2/sbstta-22-inf-04-en.pdf. Accessed 28 Sep 2021.
Halewood, M., Lopez Noriega, I., Ellis, D., Roa, C., Rouard, M., & Hamilton, R. S. (2018). Using genomic sequence information to increase conservation and sustainable use of crop diversity and benefit-sharing. Biopreservation and Biobanking. https://doi.org/10.1089/bio.2018.0043
Hartman-Scholz, A., Hillebrand, U., Freitag, J., Cancio, I., dos S. Ribeiro, C., Haringhuizen, G., Oldham, P., Saxena, D., Seitz, C., Thiele, T., & van Zimmeren, E. (2020). Finding compromise on ABS & DSI in the CBD: Requirements & policy ideas from a scientific perspective. https://www.dsmz.de/fileadmin/user_upload/Collection_allg/Final_WiLDSI_White_Paper_Oct7_2020.pdf. Accessed 28 Sep 2021.
Houssen, W., Sara, R., & Jaspars, M. (2020). Digital sequence information: Concept, scope and current use. In Digital sequence information: Concept, scope and current use. https://www.cbd.int/doc/c/fef9/2f90/70f037ccc5da885dfb293e88/dsi-ahteg-2020-01-03-en.pdf. Accessed on 28 Sep 2021.
Laird, S., & Wynberg, R. (2018). A fact-finding and scoping study on digital sequence information on genetic resources in the context of the convention on biological diversity and the Nagoya protocol. In Fact-finding and scoping study on digital sequence information on genetic resources in the context of the convention on biological diversity and the Nagoya protocol. https://www.cbd.int/doc/c/079f/2dc5/2d20217d1cdacac787524d8e/dsi-ahteg-2018-01-03-en.pdf. Accessed 28 Sep 2021.
Leonelli, S. (2019). Data – From objects to assets. Nature, 574, 317–320.
Morgera, E., Switer, S., & Geelhoed, M. (2020). Study for the European Commission on ‘Possible Ways to Address Digital Sequence Information – Legal and Policy Aspects’. https://ec.europa.eu/environment/nature/biodiversity/international/abs/pdf/Final_study_legal_and_policy_aspects.pdf. Accessed 28 Sep 2021.
Oliver, C. (1991). Strategic responses to institutional processes. The Academy of Management Review, 16, 145–179.
Randall Henning, C., & Pratt, T. (2020). Hierarchy and differentiation in international regime complexes: A theoretical framework for comparative research. https://www.peio.me/wp-content/uploads/2020/01/PEIO13_paper_66.pdf. Accessed 28 Sep 2021.
Rohden, F., Huang, S., Dröge, G., & Hartman-Sholz, A. (2020). Combined study on Digital Sequence Information (DSI) in public and private databases and traceability. In Combined study on digital sequence information in public and private databases and traceability. https://www.cbd.int/doc/c/1f8f/d793/57cb114ca40cb6468f479584/dsi-ahteg-2020-01-04-en.pdf. Accessed 28 Sep 2021.
Rohden, F., & Hartman-Sholz, A. (2021). The international political process around Digital Sequence Information under the Convention on Biological Diversity and the 2018–2020 intersessional period. Plants, People, Planet. https://doi.org/10.1002/ppp3.10198
Ruiz, M. (2015). Genetic resources as natural information: Implications for the convention on biological diversity and Nagoya protocol. Routledge.
Smyth, S. J., Macall, D. M., Phillips, P. W. B., & de Beer, J. (2020). Implications of biological information digitization: Access and benefit sharing of plant genetic resources. J World Intellect Prop, 23, 267–287. https://doi.org/10.1111/jwip.12151
Toronto International Data Release Workshop Authors. (2009). Prepublication data sharing. Nature, 461, 168–170.
Welch, E., Bagley, M., Kuiken, T., & Louafi, S. (2017). Potential implications of new synthetic biology and genomics research trajectories on the international treaty on plant genetic resources for food and agriculture. https://doi.org/10.2139/ssrn.3173781
Welch, E., Taggart, G., Feeney, M. K., & Siciliano, M. (2019). Navigating the labyrinth: Academic scientists’ responses to new regulatory controls on biological material inputs to research. Environmental Science & Policy, 110, 136–146. https://doi.org/10.1016/j.envsci.2019.08.001
Wellcome Trust. (2003). Sharing data from large-scale biological research projects: A system of tripartite responsibility. http://www.genome.gov/Pages/Research/WellcomeReport0303.pdf. Accessed 28 Sep 2021.
Wilkinson, M., Dumontier, M., Aalbersberg, I., et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci Data, 3(160018). https://doi.org/10.1038/sdata.2016.18
Disclaimer
This publication reflects the technical opinions of its authors, which are not necessarily those of the respective organizations of affiliation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2023 The Author(s)
About this chapter
Cite this chapter
Manzella, D., Marsella, M., Jaiswal, P., Arnaud, E., King, B. (2023). Digital Sequence Information and Plant Genetic Resources: Global Policy Meets Interoperability. In: Williamson, H.F., Leonelli, S. (eds) Towards Responsible Plant Data Linkage: Data Challenges for Agricultural Research and Development. Springer, Cham. https://doi.org/10.1007/978-3-031-13276-6_10
Download citation
DOI: https://doi.org/10.1007/978-3-031-13276-6_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-13275-9
Online ISBN: 978-3-031-13276-6
eBook Packages: Religion and PhilosophyPhilosophy and Religion (R0)