Abstract
Magnetic resonance imaging (MRI) has quickly established itself as the reference imaging tool for the management of patients suffering from multiple sclerosis (MS), both for the diagnosis and the follow-up of the evolution and evaluation of the impact of new therapies.
The treatment of multiple sclerosis does not cure the disease, but it slows its progression and can help to space out attacks. In this paper, tumor segmentation is treated as a problem of classification using the Ant Colony optimization algorithm (ACO) combined with a proposed protocol based on BrainSeg3D tools. Many studies and many existing approaches tend the multiple sclerosis (MS) which is a chronic inflammatory anomaly of the central nervous system.
The aim of this work is to evaluate and to verify the effectiveness of the proposed protocol on a public longitudinal database which contains 20 MS patients. This study is concerned with comparing these results against the ground truth performed by two experts and against other methods namely Dissimilarity Map (DM) creation and segmentation in terms of Dice Similarity Coefficient (DSC).
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
During the research work, we focused on brain MRI analysis, particularly in the context of monitoring the condition of patients with multiple sclerosis. MRI is considered one of the complementary examinations in the diagnostic approach of this disease since it plays a key role in monitoring the evolutionary state of the disease in patients.
The major problem lies in the difficulty of analyzing and reading these MRI images because of the variability in the size, the contrast and the difficulty of locating the lesions, and the automatic segmentation of the lesions. Several recent studies have worked on the automatic segmentation of MS lesions based on several approaches to follow the evolutionary state of this disease which can provide information on therapeutic efficacy. To detect lesions from an MRI image that carries this disease, it is necessary to adopt an efficient algorithm that can easily and efficiently detect the lesions.
To solve the problem of image segmentation and in particular to improve the performance of segmentation methods by applying them for different types of images. The objectives we have set ourselves are to contribute to the development and optimization of new segmentation criteria. We adapted the metaheuristic methods by simultaneously applying several criteria to segment the images. As we have created a new protocol based on filters and automatic tools that allow us to facilitate the task of segmentation and detection of MS lesions. Our motivation is to detect MS lesions for different patients included in novel longitudinal MRI datasets.
The residual of this study is orderly as follows: we briefly present in Sect. 2 some previous works; in Sect. 3 presents the proposed algorithm ACO_BrainSeg3D. Then we will detail the public longitudinal database we used for the test and the validation of our algorithm. Then Sect. 4 presents some results and discussion. Finally, we would end up with a conclusion and submit some perspectives.
2 Previous Works
The segmentation techniques can be classified according to different criteria and they can be presented according to a classical methodological angle. These approaches are then grouped into parametric, non-parametric and geometric approaches. Other states of the art present the different techniques by defining the classical approaches among recent approaches.
For the past twenty years, and despite the appearance of other new, more sensitive and more advanced techniques, nuclear magnetic resonance imaging has presented the tool frequently used for the diagnosis of MS since it makes it possible to detect this pathology at an earlier stage than in the past. To solve the posed problems for diagnosis, segmentation of MRI images is posed as an important task for detecting MS lesions.
In parallel with our research work for segmentation and the detection of SEP lesions, there are other similar works in progress essentially for the detection and quantification of the atrophy of the spinal cord SC in the MRI images are the key to the key. In 2018, [1] presented a fully automatic method “OPTIC” to detect the median cord line in the MRI volumes and locate the Spinal Cord (SC) in the MRI images. The authors have validated their method in healthy subjects and patients with very variable spatial resolutions and including some artifacts. The interest of this method is to detect the median line of the SC that can be used as initialization for other SC segmentation methods for pathological cases. In 2019, the authors [2] develop a fully automatic framework for segmenting SC and intra medullary lesions on a variety of MRI contrasts. The proposed approach is based on a sequence of two convening neuron networks (CNN):
-
First CNN with dilated 2D circumfolutions to detect the median line of the SC.
-
Tracking a second CNN with dilated 3D circumfolutions to segment SC and / or lesions.
In the same year, McCoy [3] used 2D convoluting neuron networks for the automatic segmentation of the SC and traumatic axial MRI contusion in T2 in a cohort of patients with acute SC injury. They have developed a image analysis pipeline integrating 2D convolutionary neuron networks for the segmentation of the entire and intramedullary spinal cord lesions.
In Article of [4] to segment the lesions in the pathology of multiple sclerosis, they studied two stages of pretreatment based on skull stripping (SS) and the improvement of the contrast which are two important steps to improve the quality rate of MS lesion segmentation. After the pretreatment step, a segmentation approach based on the method of maximizing expectations (EM) has been applied to extract SEP lesions. Again study of [5] for the contrast improvement of MRI images for a better appearance of normal tissues and sick tissues affected by the pathology of MS.
The important recent segmentation study presents a new protocol to develop benchmark segmentations of white matter tumors based on multi-rater consensus using a new MR database of 30 patients with multiple sclerosis [6]. They have developed advanced BrainSeg3Dsoftware which allows their correct and effective delineation in 3D MR images.
From this study, we carried out work based on the tools of the BrainSeg3D software and this new database: the first work is to create a novel protocol applied the tools of the BrainSeg3D software applied on this new database which contains 30 MS patients.
The new database is tested by the metaheuristic ant colony algorithm used by [7, 8] helping to improve the parameters of this algorithm [9, 10].
The research [10] manages to realize a new ACO_BrainSeg3D segmentation algorithm that combines the ACO algorithm and the protocol based on BrainSeg3D tools, this approach is tested on the novel longitudinal dataset with reference.
Another recent study [11] objectively validate and compare several intensity-based methods for detecting white matter tumor changes using longitudinal MR image datasets with accurate ground truth.
The Validation of change detection focused on evaluating the two main steps of change detection, namely Dissimilarity Map (DM) creation and segmentation. Therefore, combinations of three methods for DM creation (Subtraction of intensity images (STI), Generalized Like Lihood Ratio (GLR) and Logistic Regression Model (LRM)) and three for DM segmentation (Confidence level thresholding (CLT), Change vector angular histogram thresholding (CVAHT) and optimal threshold) were tested on 20 MR image datasets with a common pre-processing and post-processing.
It was created by two specialist evaluators who subtracted and segmented of white matter tumor of two studies: baseline and follow-up T1w, T2w and FLAIR MR Images. The reference segmentations were cooperatively treated by the evaluators till they heard on what was inspected the reference segmentation. This publicly dataset and the BrainSeg3D software are available on site web http://lit.fe.uni-lj.si/tools.
In this study, the objective is to employ the proposed algorithm ACO_BrainSeg3D on a new longitudinal dataset containing 20 MS patients in order to evaluate the performance and reliability of this algorithm against the ground truth and other tested methods on the same database.
The aim of the actual work was to supply a literal test and to compare the results of propound algorithm with the three intensity based approaches Dissimilarity Map creation and segmentation combinations (STI/CLT, STI/CVAHT and STI/Optimal threshold), (GLR/CLT, GLR/CVAHT and GLR/Optimal threshold) and (LRM/CLT, LRM/CVAHT and LRM/Optimal threshold) [11].
We can thus note that almost all the outliers are identified by the ACO_BrainSeg3D protocol in front of the reference, and compared to three methods for DM creation (STI, GLR and LRM) combined with three for DM segmentation (CLT, CVAHT and optimal threshold) in terms of the median coefficient of similarity.
We have tested the proposed algorithm using public longitudinal available dataset with brain MR Images containing two studies: baseline and Follow-up studies with three MRI sequences T1-w, T2-w and Flair. In all cases, our algorithm outperformed competing approaches. The obtained evaluations reported good results, where all the outliers have emerged in spite of there being falsely spotted outliers.
3 The Proposed ACO_BrainSeg3D Algorithm
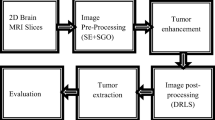
In this paper we expose the suggested method which used the combination of ACO algorithm with a propound protocol by using existing and new semi-automated outliers segmentation tools of BrainSeg3D software. So as to facilitate the tumors segmentation, we employed the MATLAB software and the specialized BrainSeg3D software. Our proposed contribution is named ACO_BrainSeg3D.
The global function of the propound contribution is detailed on Fig. 1.
The proposed algorithm is essentially based on two algorithms as detailed below:
All MRI images from the database used are first processed by the ACO metaheuristic algorithm. Also in the studies [9, 12, 13] which are based on ACO algorithm with the same database, when we obtained good results compared to reference of the novel MR Datasets.
Concerning this algorithm, our contribution is mainly focused on the variation of the parameters empirically in order to have good results for the segmentation of the images of the new longitudinal database and a good detection of MS lesions. The latter consists of three parameters to better characterize good quality segmentation. Then, the segmentation thresholds maximizing the criteria are sought using the ACO algorithm.
The settings and the essential equations of ACO algorithm are detailed in our studies [9, 10]. Then the major tested values to do all simulations are presented in table 1.
Our important contribution is the testing of parameters in order to obtain good results. This task is done in [12, 13] and it evaluated according to existing research and the consensus of segmentation.
4 Validation Dataset
The validated dataset contains two studies baseline and follow-up MR images for 20 subjects with multiple sclerosis as illustrated in Fig. 2.
The images were acquired on a 1.5 T Philips MRI machine at the University Medical Centre Ljubljana (UMCL).
Every subjects of this dataset contains a 2D T1-weighted, 2D T2-weighted and a 2D FLAIR image with different setting indicated in the Table 2.
The reference of MR images was proposed by two expert evaluators for evaluation purposes. The 20 MS patients with baseline and follow-up studies with respectively T1-W, T2-W and Flair images are available in the site http://lit.fe.uni-lj.si/tools.
5 Results and Discussion
In order to validate the capability of our algorithm to detect MS tumor, we calculated three essential terms of validation: the coefficient DSC, TPR (True positive Rate) and robustness of FPR (False Positive Rate).
The True Positive Rate corresponds to sensitivity:
The False Positive Rate corresponds to precision that explains the capability to select all existing outliers corresponds to the value:
The Dice Similarity Coefficient also corresponds to the Kappa Index (or Dice coefficient) and measures the overlap between the test of segmentation obtained and the consensus. The Dice Similarity Coefficient (DSC) [15] can therefore be calculated with respect to consensus segmentation:
Our important goal is to detect all outliers, True positive (TP) presents a detected region if at least one voxel overlaps the reference, otherwise it is considered a false positive (FP). Any labeling not spotted by our approach is considered as a false negative (FN) [16]. Whose values fluctuate in [0, 1] and a value superior 0.6 presented a good conformity with the ground truth.
We see here in Fig. 5 that the proposed algorithm manages to detect almost all the lesions in the axial slices of the original image as for patients 1, 3, 6, 8 and 11. For patients 7 and 19, there are slightly visible lesions not detected. And for the other patients, they are considered as healthy patients due to the absence of lesions; we can only discover this for a single patient 17 in which the proposed algorithm detects a small, slightly visible lesion.
The results of segmentation by the suggested algorithmACO_BrainSeg3D are exposed in Fig. 5. This algorithm yields good results, where all the outliers have arisen in spite of there being falsely spotted lesions.
The respective values procured with our proposed ACO_BrainSeg3D algorithm of DSC are between 0.44 and 1, the values of FPR are in the interval [0.5 1] and the values of TPR are between 0.33 and 1(Fig. 3). We mark that the validation values obtained in this test are lightly elevated than these reported on every outliers segmentation publicized in the literature [11].
We managed in this paper to detect several lesions according to the number and volume in the first phase of advancement of the lesions it is the relapsing remittent form RR for all the patients of the database with a percentage of 70% of rate of advancement, as we detected lesions in the secondary progressive form SP with a portion of 5% and a percentage of 25% of other patients not specified or we cannot identify whether these patients are really healthy or there are very small lesions in the early phase that we couldn't detect (Fig. 4).
This analysis has been processed for some patients in the database for example for the axial sections of the flair image for patients 2, 4, 5, 20 … as illustrated in Fig. 5.
This is due to the fact that we can see the positions of further outliers in link with the consensus, and we can spot the missed outliers, which could otherwise have a strong impact on DSC.
We objectively evaluated in this paper the proposed algorithm which combine ACO algorithm with the protocol using the BrainSeg3D tools on longitudinal MRI database of 20 MS subjects.
The aim of the actual work was to supply a literal test and to compare the results of propound algorithm with the three intensity based approaches Dissimilarity Map creation and segmentation combinations (STI/CLT, STI/CVAHT and STI/Optimal threshold), (GLR/CLT, GLR/CVAHT and GLR/Optimal threshold) and (LRM/CLT, LRM/CVAHT and LRM/Optimal threshold).
To explain the impact on the median DSC, Table 3 reports corresponding median values of the proposed algorithm and the combinations methods based on segmentation and creation DM after the post-processing.
The median DSC after post-processing of three combinations of three change segmentations DM and three creation DM approaches was consistently reduced in accordance of our suggested algorithm where the value DSC is 0.76.
The middle DSC value for the ACO_BrainSeg3D algorithm is studied as follows:
Aside from various fusions of the DM creation and segmentation approaches were tested by using the same quantitative metrics. Comparative test according to three DM alternatives (Table 3) illustrated an elevated average DSC (76%).
This result reports that the used outliers segmentation algorithm marked very substantial and reliable tumor segmentations.
That our segmentation algorithm of segmentation applied on the novel longitudinal database is significantly higher compared to the DSC of DM creation combined with three DM segmentation. So the ACO_BrainSeg3D algorithm detects more lesions compared to ground truth.
The advantage of proposed algorithm ACO_BrainSeg3D applied on this longitudinal database is the detection of almost all existing lesions in the ground truth thanks to the values carried out within the acceptable measurement interval. Our algorithm needs more run time but we don't have a real time constraint. The only drawback is the non-detection of small lesions which are not very visible, which encourages the researchers to improve their techniques until they detect small lesions of a small volume.
We can thus note that almost all the outliers are identified by the ACO_BrainSeg3D protocol in front of the reference, and compared to three methods for DM creation (STI, GLR and LRM) combined with three for DM segmentation (CLT, CVAHT and optimal threshold) in terms of the median coefficient of similarity.
This validation is carried out first based on the DSC coefficient largely which indicates similarity and second concerning the ability and precision to detect the number of tumors existing in the ground truth based on the TPR and the FPR.
In order to mark this object and allow other researchers to replicate the test of this work, we wish to extend and publicly propagate the encouraging results of our proposed algorithm using the longitudinal database available on the site: http://lit.fe.uni-lj.si/tools.
We consider this study as the first but important step towards the realization of a protocol based on BrainSeg3D software tools combined with an improved metaheuristic algorithm applied to different public databases for validation and comparison of detection methods with longitudinal and other changes.
6 Conclusion
In this paper, we propounded an automatic protocol for cerebral tumors segmentation in longitudinal MRI images. The idea was evaluated the ACO algorithm by using the MATLAB software and protocol based on semi-automated tools using BrainSeg3Dsoftware with a for longitudinal MRI image dataset which contains 20 patients with multiple sclerosis with reference agreed by two domain specialist.
We have tested the proposed algorithm using public longitudinal available dataset with brain MR Images containing two studies: baseline and Follow-up studies with three MRI sequences T1-w, T2-w and Flair. In all cases, our algorithm outperformed competing approaches. The obtained evaluations reported good results, where all the outliers have emerged in spite of there being falsely spotted outliers.
A short-term perspective is to make an interface that implements our proposed algorithm ACO_Brainseg3D on computer. This can facilitate the use of the method and gain more time. It is necessary to test this interface on larger databases to clear the strengths and limitations. This step is essential before exporting our algorithm in a clinical context.
References
Gros, C., et al.: Automatic spinal cord localization, robust to MRI contrasts using global curve optimization. Med. Image Anal. (2017)
Gros, C., et al.: Automatic segmentation of the spinalcord and intramedullary multiple sclerosis lesions with convolutionalneural networks. Neuroimage 184, 901–915 (2019)
McCoy, D.B., et al.: Convolutional neural network-based automated segmentation of the spinal cord and contusion injury: deep learning biomarker correlates of motor impairment in acute spinal cord injury. Am. J. Neuroradiol. 40, 737–744 (2019)
Sahnoun, M., et al.: Contrast-enhanced image analysis for MRI based multiple sclerosis lesion segmentation. In: 5th International Conference on Advanced Technologies For Signal and Image Processing, ATSIP 2020, Sfax, Tunisia, 2–5 Sept. 2020
Sahnoun, M., Kallel, F., Dammak, M., Mhiri, C., Ben Mahfoudh, K., Ben Hamida, A.: A comparative study of MRI contrast enhancement techniques based on traditional gamma correction and adaptive gamma correction: case of multiple sclerosis pathology. In: The 4th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP) (2018)
Lesjak, Ž, et al.: A novel public MR image dataset of multiple sclerosis patients with lesion segmentations based on multi-rater consensus. Neuroinformatics 16(1), 51–63 (2017). https://doi.org/10.1007/s12021-017-9348-7
Dorigo, M., Blum, C.: Ant colony optimization theory: a survey. Theor. Comput. Sci. 344, 243–278 (2005)
Devers, L., Fauvel, L.: TER MIASHS 2017. Université Bordeaux
Bouzidi, D., Ghozzi, F., Fakhfakh, A.: Ant colony optimisation ACO to detect multiple sclerosis lesions using magnetic resonance imaging. In: Bouktir, T., Neji, R. (eds.) Proceedings of the 4th International Conference on Recent Advances in Electrical Systems, pp. 73–77. Tunisia (Dec 2019). ISBN: 978-9938-9937-2-1
Bouzidi, D., Ghozzi, F., Taouil, K., Fakhfakh, A.: MR brain image segmentation optimized by using ant colony algorithm with BrainSeg3D for multiple sclerosis tumors detection. NAUN Int. J. Biol. Biomed. Eng., 218–228. https://doi.org/10.46300/91011.2020.14.29
Lesjak, Ž, Pernuš, F., Likar, B., Špiclin, Ž: Validation of white-matter lesion change detection methods on a novel publicly available MRI image database. Neuroinformatics 14(4), 403–420 (2016). https://doi.org/10.1007/s12021-016-9301-1
Bouzidi, D., Ghozzi, F., Fakhfakh, A.: MR brain image segmentation optimized by using ant colony algorithm. In: 21th International Multi-Conference on Systems, Signals Devices (SSD)
Bouzidi, D., Ghozzi, F., Fakhfakh, A.: Ant colony optimization algorithm for multiple sclerosis lesion segmentation based on MR brain image. Advances in Communication, Signal and Image Processing (2022) (in press)
Bouzidi, D., Ghozzi, F., Taouil, K., Fakhfakh, A.: BrainSeg3D to detect Multiple sclerosis lesions using magnetic resonance imaging. In: The IEEE International Conference on Design and Test of Integrated Micro and Nano-Systems. IEEE, Tunisia, June 2020. https://doi.org/10.1109/DTS48731.2020.9196053. ISBN: 978-1-7281-5429- 9/20/$31.00 ©2020
Dice, L.R.: Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945)
Zijdenbos, A.P., Forghani, R., Evans, A.C.: Automatic pipeline analysis of 3-D MRI data for clinical trials: application to multiple sclerosis. IEEE Trans. Med. Imag. 21, 1280–1291 (2002)
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this paper
Cite this paper
Bouzidi, D., Ghozzi, F., Fakhfakh, A. (2022). Ant Colony Optimization with BrainSeg3D Protocol for Multiple Sclerosis Lesion Detection. In: Aloulou, H., Abdulrazak, B., de Marassé-Enouf, A., Mokhtari, M. (eds) Participative Urban Health and Healthy Aging in the Age of AI. ICOST 2022. Lecture Notes in Computer Science, vol 13287. Springer, Cham. https://doi.org/10.1007/978-3-031-09593-1_19
Download citation
DOI: https://doi.org/10.1007/978-3-031-09593-1_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-09592-4
Online ISBN: 978-3-031-09593-1
eBook Packages: Computer ScienceComputer Science (R0)