Abstract
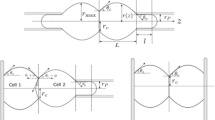
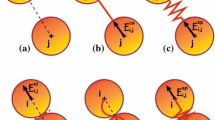
This work introduces a computational model of elastic double cluster. We describe a method to create a partially flattened spherical cell and a mirroring process that creates a symmetrical double cluster with desired adhesion surface. The main focus is on the adhesion between the two cells modeled by repulsive-attractive Lennard-Jones potential. We study the stability of the adhesion with respect to the parameters of the Lennard-Jones potential and to the elasticity of the cells. Based on these, a baseline cluster is created and calibrated to a specific separation force using computational experiment that mimics a dual micropipette assay. This cluster is then immersed into elongation flow to create a parallel between the two types of cell stretching experiments: one that mechanically pulls the cell membrane and another where fluid flow creates stress on the membrane. Thus validated, our model of adhesion can be used in more complex clusters and serve as a building block in future computational studies.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Anderson, K.J., de Guillebon, A., Hughes, A.D., Wang, W., King, M.R.: Effect of circulating tumor cell aggregate configuration on hemodynamic transport and wall contact. Math. Biosci. 294, 181–194 (2017)
Au, S.H., et al.: Clusters of circulating tumor cells traverse capillary-sized vessels. Proc. Natl. Acad. Sci. 113(18), 4947–4952 (2016)
Bithi, S.S., Vanapalli, S.A.: Microfluidic cell isolation technology for drug testing of single tumor cells and their clusters. Sci. Rep. 7(1), 1–12 (2017)
Bonnet, J., et al.: Mitotic arrest affects clustering of tumor cells. Cell Div. 16(1), 1–13 (2021)
Choi, J.W., Kim, J.K., Yang, Y.J., Kim, P., Yoon, K.H., Yun, S.H.: Urokinase exerts antimetastatic effects by dissociating clusters of circulating tumor cells. Can. Res. 75(21), 4474–4482 (2015)
Cimrák, I., Jančigová, I.: Computational Blood Cell Mechanics, 1st edn. CRC Press, Boca Raton (2018)
Dabagh, M., Gounley, J., Randles, A.: Localization of rolling and firm-adhesive interactions between circulating tumor cells and the microvasculature wall. Cell. Mol. Bioeng. 13(2), 141–154 (2020)
Daoudi, M., et al.: Enhanced adhesive capacities of the naturally occurring ile249-met280 variant of the chemokine receptor cx3cr1. J. Biol. Chem. 279(19), 19649–57 (2004)
Edd, J.F., et al.: Microfluidic concentration and separation of circulating tumor cell clusters from large blood volumes. Lab Chip 20(3), 558–567 (2020)
Hochmuth, R.M.: Micropipette aspiration of living cells. J. Biomech. 33(1), 15–22 (2000)
Jančigová, I., Kovalčíková, K., Bohiniková, A., Cimrák, I.: Spring-network model of red blood cell: From membrane mechanics to validation. Int. J. Numer. Meth. Fluids 92(10), 1368–1393 (2020)
Jančigová, I., Kovalčíková, K., Weeber, R., Cimrák, I.: Pyoif: computational tool for modelling of multi-cell flows in complex geometries. PLOS Comput. Biol. 16(10), 1–21 (2020)
King, M.R., et al.: A physical sciences network characterization of circulating tumor cell aggregate transport. Am. J. Physiol. Cell Physiol. 308(10), C792–C802 (2015)
Kovalcikova, K., Bachraty, H., Bachrata, K., Buzakova, K.: Numerical experiment characteristics dependence on red blood cell parameters, pp. 286–292 (2021)
Maître, J.L., et al.: Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science 338(6104), 253–256 (2012)
Marrella, A., et al.: High blood flow shear stress values are associated with circulating tumor cells cluster disaggregation in a multi-channel microfluidic device. PLoS ONE 16(1), e0245536 (2021)
Mutlu, B.R., et al.: In-flow measurement of cell-cell adhesion using oscillatory inertial microfluidics. Lab Chip 20(9), 1612–1620 (2020)
PyOIF: Computational Tool for Modelling of Multi-Cell Flows in Complex Geometries: Resources webpage for pyoif. online January 2022. https://cellinfluidfriunizask/resources-espresso/
Rapaport, D.C.: The Art of Molecular Dynamics Simulation, 2nd edn. Cambridge University Press, Cambridge (2004)
Shaw Bagnall, J., Byun, S., Begum, S.E.A.: Deformability of tumor cells versus blood cells. Sci Rep 5(1), 1–11 (2015)
TruongVo, T., et al.: Microfluidic channel for characterizing normal and breast cancer cells. J. Micromech. Microeng. 27(3), 035017 (2017)
Acknowledgements
This research was supported by Operational Program “Integrated Infrastructure” of the project “Integrated strategy in the development of personalized medicine of selected malignant tumor diseases and its impact on life quality”, ITMS code: 313011V446, co-financed by resources of European Regional Development Fund.
This work was also supported by the Slovak Research and Development Agency (contract number APVV-15-0751).
James J. Feng acknowledges support by the Natural Sciences and Engineering Research Council of Canada (Discovery Grant No. 2019-04162).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 Springer Nature Switzerland AG
About this paper
Cite this paper
Bohiniková, A., Jančigová, I., Cimrák, I., Feng, J.J. (2022). Sensitivity Analysis of Adhesion in Computational Model of Elastic Doublet. In: Rojas, I., Valenzuela, O., Rojas, F., Herrera, L.J., Ortuño, F. (eds) Bioinformatics and Biomedical Engineering. IWBBIO 2022. Lecture Notes in Computer Science(), vol 13347. Springer, Cham. https://doi.org/10.1007/978-3-031-07802-6_19
Download citation
DOI: https://doi.org/10.1007/978-3-031-07802-6_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-07801-9
Online ISBN: 978-3-031-07802-6
eBook Packages: Computer ScienceComputer Science (R0)