Abstract
A wealth of public data repositories is available to drive genomics and clinical research. However, there is no agreement among the various data formats and models; in the common practice, data sources are accessed one by one, learning their specific descriptions with tedious efforts. In this context, the integration of genomic data and of their describing metadata becomes—at the same time—an important, difficult, and well-recognized challenge. In this chapter, after overviewing the most important human genomic data players, we propose a conceptual model of metadata and an extended architecture for integrating datasets, retrieved from a variety of data sources, based upon a structured transformation process; we then describe a user-friendly search system providing access to the resulting consolidated repository, enriched by a multi-ontology knowledge base. Inspired by our work on genomic data integration, during the COVID-19 pandemic outbreak we successfully re-applied the previously proposed model-build-search paradigm, building on the analogies among the human and viral genomics domains. The availability of conceptual models, related databases, and search systems for both humans and viruses will provide important opportunities for research, especially if virus data will be connected to its host, provider of genomic and phenotype information.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
1 Introduction
Genomics was born in relatively recent times: in the last two decades, after the introduction of Next Generation Sequencing (NGS) technologies [27], the processes of DNA/RNA sequencing have benefit form notable cost and time reductions. NGS data is employed at three levels. Primary analysis produces raw datasets of nucleotide bases, reaching a typical size of 200 Gigabytes per single human genome when stored [30]. Secondary analysis produces regions of interest, including mutations (where the code of an individual differs from the code of the “reference” human being) – possibly associated with genetic diseases and cancers, gene expression (indicating in which conditions genes are active), and epigenomic signals (phenotype changes not involving alterations in the genetic sequence). Finally, tertiary analysis aggregates and combines together heterogeneous datasets produced during the preceding phase, trying to “making sense” of the data, unveiling complex biological mechanisms.
For boosting the last—and most interesting—type of analysis, thousands of datasets are becoming available every day, typically produced within the scope of large cooperative efforts, open for public use and made available for secondary research use [13], including the Encyclopedia of DNA Elements (ENCODE, [22]), Genomic Data Commons (GDC, [23]), Gene Expression Omnibus (GEO, [3]) Roadmap Epigenomics [24], and the 1000 Genomes Project [1]. In addition to these well-known sources, we are witnessing the birth of several initiatives of population-specific or nation-scale sequencing [29].
In the following, we focus on processed genomic datasets, which include experimental observations—representing regions along the genome chromosomes with their properties—and metadata, carrying information about the observed biological phenomena. The integration of genomic data and of their describing metadata is a challenge that is at the same time important (as a wealth of public data repositories is available to drive biological and clinical research), difficult (as the domain is complex and there is no agreement among the various data formats and definitions), and well-recognized (because, in the common practice, repositories are accessed one-by-one, with tedious and error-prone efforts). Although the potential collective amount of available information is huge, the effective combination of genomic datasets is hindered by their heterogeneity (in terms of download protocols, formats, notations, attribute names, and values) and lack of interconnectednes.
Motivating Example Let us consider a researcher who is looking for data to perform a comparison study between a human non-healthy breast tissue, affected by carcinoma, and a healthy sample coming from the same tissue type. Exploiting her previous experience, the researcher locates three portals having interesting data for this analysis (see Fig. 1). On GDC, one or more cases can be retrieved with the query “disease = Breast Invasive Carcinoma”. To compare such data with references, the researcher chooses additional datasets coming from cell lines, a standard benchmark for investigations. On the GEO web interface, tumor cell line data is found by browsing thousands of human samples (e.g., the “T47D-MTVL” exhibits the disease “breast cancer ductal carcinoma”). Finally, on ENCODE, the researcher chooses both a tumor cell line (“MCF-7”, affected by “Breast cancer (adenocarcinoma)”) and a normal cell line(“MCF-10A”, widely considered the non-tumorigenic counterpart), to make a control comparison. As it can be noted, from both points of view of attributes and of values—when searching for disease-related information—we find many forms, only possibly pointing to comparable samples. This kind of information is not encoded in a unique way over data sources and is often missing. Considerable external knowledge is necessary to find appropriate connections; this cannot be obtained on the mentioned portals, but needs to be retrieved manually by querying specific databases, dedicated forums, or specialized ontologies.
In this chapter we describe the research carried out in the context of the Genomic Computing ERC project [20], concerned with designing and building a repository of processed NGS data genomic datasets using a systematic and repeatable approach:
-
Model: we analyze the domain state of the art (including scouting of online resources/documentation and testing data retrieval methods). Data is studied with the goal of proposing a conceptual model for the main characteristics shared by relevant data sources in the field, targeting completeness but favoring simplicity, for producing easy-to-use systems for biologists and genomic experts.
-
Integrate and build: we select interesting open data sources for the domain, build solid pipelines to download data from them, and transform it into a standard interoperable format, obtaining a repository of homogenized data, to be used seamlessly from a unique endpoint, allowing integrative biological queries.
-
Search: we target the end-users of the repository, i.e., experts of the domain who browse the repository in search for datasets to prove or disprove their research hypotheses. Interfaces need to take into account their background: considerable biological knowledge, but limited understanding of programming languages.
During the first phase of the COVID-19 epidemic, in March and April 2020, we responded proactively to the call to arms issued by the broad scientific community. We conducted an extensive requirement analysis by engaging in interdisciplinary conversations with a variety of scientists, including virologists, geneticists, biologists, and clinicians. This preliminary activity convinced us of the need for a structured proposal for viral data modeling and management. We thus reapplied the previously proposed methodology of modeling a data domain, integrating many sources to build a global repository, and finally making its content searchable to enable further analysis. This experience suggests that our approach is general enough to be applied to any domain of life sciences and encourages broader adoption.
Chapter organization. Section 2 describes our data integration proposal in the field of genomic tertiary analysis; Sect. 3 is dedicated to understanding the world of viral sequences and their descriptions (collected samples, host characteristics, variants and their impact on the related disease); Sect. 4 foresees how the two previous sections could be included in one single system to drive powerful biological discovery.
2 Human Genomic Data Integration
By analysing the players involved in the genomic data context [13], we proposed the Genomic Conceptual Model (GCM, [7]), which captures the most common metadata attributes that describe genomic data items and experiments in the available sources. The model (drawn in Fig. 2 represents a typical genomic region data file (i.e., the Item), using different perspectives: the biology (including information on the donor, the derived biological samples and different experimental replicates), the technology (with assay details and observed features), the management and organization aspects of the experiment, and the extraction parameters. GCM sets the basis for querying the underlying data sources for locating relevant experimental datasets.
We formalized a methodology for integrating and making datasets interoperable, called META-BASE architecture [4, 11], which is focused on obtaining a usable and consistent result, also from the data quality point of view [5]. As shown in Fig. 3, META-BASE takes care of retrieving datasets as partitions of a number of relevant data sources, transforming them into semi-structured datasets with an interoperable format (GDM [25]), cleaning redundant information, and joining their schemata into a global view represented by the GCM logical and physical implementation into a relational database. Later, we apply a value-normalization and enrichment procedure that exploits the content of several curated biomedical ontologies (such as OBI, NCIT, UBERON) for pointing to concepts (and their hierarchies) that are well-known in the domain [8]. The pipeline is general, open and extensible, being able to easily incorporate any number of new sources.
The resulting repository—already integrating several important sources such as ENCODE, The Cancer Genome Atlas from GCC [19] (via the OpenGDC framework), Roadmap Epigenomics, and the 1000 Genomes Project, contains now about 560K items, grouped within 67 datasets, collectively occupying more than 9TB of memory. A noteworthy metadata extraction framework has been implemented for the GEO source [18], where we employ a transformer-based machine learning approach to gather structured metadata from the experiments’ textual descriptions.
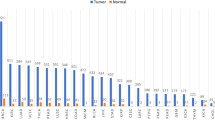
The repository is exposed by means of user interfaces to respond to biological researchers’ needs. We provide two different interfaces. The first one is a graph-based endpoint for expert users, who are focused on understanding the inference process performed on metadata in order match items in the repository [10]. Thanks to the ontological enrichment of metadata, we allow for a powerful semantic search mechanism. Consider, for example, choosing the tissue “Uterus”. As shown in Fig. 4, this concept subsumes several other terms, which can be matched when different search levels are selected. In Table 1 we show the number of genomic items (i.e., region data files) that can be retrieved by using the Orig. level (only matching items described by exact keywords), Syn. level (also matching items described by synonyms and alternative forms), or Exp. level (also matching the sub-concepts of “uterus”), respectively resulting into the retrieval of 57, 1708, or 16851 items.
The second interface, GenoSurf [16] (http://gmql.eu/genosurf/) is a user-friendly search system providing access to the consolidated repository of metadata attributes, also enriched by a multi-ontology knowledge base, locating relevant genomic datasets, which can then be analyzed with off-the-shelf bioinformatic tools. The section of the Data Search interface of GenoSurf results from the translation of the GCM model into a much simpler denormalized structure consisting of a star with four related dimensions. This interface was evaluated by running an extended empirical study whose participants were knowledgeable in Biology and Computer Science. We collected many relevant insights related to the data scouting and extraction habits of different user profiles [9].
The frameworks and tools described in this section are included in a follow-up project, to be exploited for providing biologists and clinicians with a complete data extraction/analysis environment [21] that is: (i) guided by a conversational interface; (ii) equipped with a “marketplace” of ready-to-use best practices; we thus aim to break down the technological barriers that are currently hindering the practical adoption of our systems.
3 Virus Sequence Data Integration
Inspired by our work on genomic data integration, during the outbreak of the COVID-19 pandemic we searched for effective ways to help mitigate its effects with our contribution. As a first step, we conducted several interviews to experts and candidate users of our perspective systems [6], to quickly build the necessary expertise to operate in this new field. We understood that the collection of viral genome sequences is of paramount importance, in order to study the origin, wide spreading and evolution of SARS-CoV-2 (the virus responsible for the COVID-19 disease). Since the beginning of the pandemic, we have observed an almost exponential growth of the number of sequences deposited to known databases [14]: from few hundreds in March 2020, to one hundred thousand in August 2020, to almost two millions as of June 2021. Note that this is the first time that NGS technologies are been used for sequencing a massive amount of viral sequences. In several cases, also relevant associated data and metadata are provided, although their amount, coverage and harmonization are still limited.
Several institutions provide databases and resources for depositing viral sequences. Some of them, such as NCBI’s GenBank [26], preexist the COVID-19 pandemic, as they host thousands of viral species – including, e.g., Ebola, SARS and Dengue. Other organizations have produced new data collections specifically dedicated to the hosting of SARS-CoV-2 sequences. It is the case of COG-UK [31], a pioneering project in the United Kingdom that has produced about one fourth of world-wide SARS-CoV-2 sequuences, and GISAID [28], which has soon become the worldwide predominant data source. While GenBank and COG-UK have adopted a fully open-source model of data distribution and sharing, GISAID is protecting the deposited sequences by having users login from an institutional site and accept a Database Access Agreement. We decided to re-apply the model-build-search paradigm used for human genomics. Building integrated databases for viral genomics and related search systems for accessing them is of uttermost importance for controlling the current pandemic and future ones. Several molecular biology studies can then be supported, considering haplotypes (i.e., clusters of inherited variations at single positions genomic sequence), phylogenetic trees (i.e., diagrams for representing the evolutionary relationships among organisms), and the evolution of new variants in general. Unfortunately, several limitations are still present, including the impact of GISAID’s model, the lack of metadata quality, and the (un)willingness of sequence sharing, especially in some countries.
Understanding viruses from a conceptual modeling perspective is very important. Even if the domain of viral genomics is completely new, it presents many analogies with our previous challenges. Here, we model viral nucleotide sequences as strings of letters, with corresponding sub-sequences – the genes – that encode for amino acid proteins [12]. In our Viral Conceptual Model (Fig. 5) the Sequence of the virus is the central information. We describe a sequence’s aspects using four perspectives: (i) technological (about the sequencing experiment); (ii) biological (about the virus—belonging to a complex taxonomy—isolated from an infected host and the isolation source from which viral material is extracted); (iii) organizational (the sequencing project, the hosting databases, the scientific and medical publications related to the discovery of sequences); (iv) analytical (the sequence’s annotated parts—known genes, coding and untranslated regions—and the nucleotide/amino acids variants, computed with respect to the reference sequence of the species).
We then integrate sequences with their metadata from a variety of different sources. When stored into a unique database, virus strains may be searched and compared intra- and cross-species. We propose the powerful search interface ViruSurf [17] (http://gmql.eu/virusurf/), able to quickly extract sequences based on their combined variants, to compare different conditions, and to build interesting populations for downstream analysis. When applied to SARS-CoV-2, the virus responsible for COVID-19, complex conceptual queries upon our system are able to replicate the search results of recent articles, hence demonstrating considerable potential in supporting virology research. ViruSurf has being extended to accommodate other types of data, e.g., epitopes, which are sub-sequences of amino acids from a virus protein antigen that can activate an immune response from the host, being relevant for vaccine design. We produced EpiSurf (http://gmql.eu/episurf/, https://doi.org/10.1093/database/baab059), a web server for selecting viral populations of interest and analyzing how their amino acid changes are distributed along epitopes. In addition, we have provisioned the two search servers with the visual and analytical support by VirusViz [15] (http://gmql.eu/virusviz/); the application allows to show distributions of nucleotide and amino acid mutations, build groups (i.e., sequence populations of interest) and to compare their variation distributions. VirusViz provides examples related to SARS-CoV-2 variants of concern/interest (initially observed in UK, California, and New York), demonstrating how new variants can be traced since their starting dates (see https://github.com/DEIB-GECO/VirusViz/wiki/Supplementary-material-examples). VirusViz is also enriched by a knowledge base of amino acid changes effects (spanning from protein stability to epidemiological/immunological aspects), comprising for example viral transmission, fitness, binding affinity to host receptor and sensitivity to specific treatments (see CoV2K, [2]).
4 Conclusions
In Sect. 2, we presented our approach to modeling human genomic data, building a sound repository of genomic datasets using data integration techniques, and exposing its content through user interfaces that are rich in functionalities and data complexity. Our commitment is to continue the inclusion of relevant data sources for bioinformatics tertiary analysis, improving our process from a data quality and interoperability point of view. In Sect. 3, we have presented our approach to modeling viral sequences, building a repository collecting data from different viral data sources, and exposing its content over a first web interface, with enhanced functionalities on variant selection and filtering. This work has been realized in the first nine months of the SARS-CoV-2 epidemic worldwide. After setting the first milestones, we have already moved forward, considering the next challenges of this new domain with growing interest. More in general, the results on this thesis are part of a broad vision: availability of conceptual models, related databases and search systems for both human and viral genomics will provide important opportunities for genomic and clinical research, especially if virus (or other pathogens) sequences can be connected to the genotype and phenotype information regarding its host, i.e., the human organism, as suggested by Fig. 6. Such integration would drive more powerful biological discovery, being of particular interest for future epidemics events.
References
1000 Genomes Project Consortium, A global reference for human genetic variation. Nature 526(7571), 68 (2015)
R. Al Khalaf, T. Alfonsi et al., CoV2K: A Knowledge Base of SARS-CoV-2 Variant Impacts, in Research Challenges in Information Science (RCIS 2021) (Springer, Cham, 2021)
T. Barrett, S.E. Wilhite et al., NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 41(D1), D991–D995 (2012)
A. Bernasconi, Using metadata for locating genomic datasets on a global scale, in Data and Text Mining in Biomedical Informatics (DTMBio 2018), CEUR Workshop Proceedings, vol. 2482 (2018)
A. Bernasconi, Data quality-aware genomic data integration. Comput. Meth. Prog. Biomed. Update 1, 100009 (2021)
A. Bernasconi, Extreme requirements elicitation: lessons learnt from the COVID-19 case study, in Requirements Engineering: Foundation for Software Quality (REFSQ 2021), CEUR Workshop Proceedings, vol. 2857 (2021)
A. Bernasconi, S. Ceri et al., Conceptual Modeling for Genomics: Building an Integrated Repository of Open Data, in Conceptual Modeling (ER 2017) (Springer, Cham, 2017), pp. 325–339
A. Bernasconi, A. Canakoglu et al., Ontology-driven metadata enrichment for genomic datasets, in Semantic Web Applications and Tools for Life Sciences (SWAT4LS 2018), CEUR Workshop Proceedings, vol. 2275 (2018)
A. Bernasconi, A. Canakoglu, S. Ceri, Exploiting conceptual modeling for searching genomic metadata: a quantitative and qualitative empirical study, in Advances in Conceptual Modeling (EmpER 2019) (Springer, Cham, 2019), pp. 83–94
A. Bernasconi, A. Canakoglu, S. Ceri, From a conceptual model to a knowledge graph for genomic datasets, in Conceptual Modeling (ER 2019) (Springer, Cham, 2019), pp. 352–360
A. Bernasconi, A. Canakoglu et al., META-BASE: a novel architecture for large-scale genomic metadata integration. IEEE/ACM Trans. Comput. Biol. Bioinf. (2020)
A. Bernasconi, A. Canakoglu et al., Empowering Virus Sequence Research through Conceptual Modeling, in Conceptual Modeling (ER 2020) (Springer, Cham, 2020), pp. 388–402
A. Bernasconi, A. Canakoglu et al., The road towards data integration in human genomics: players, steps and interactions. Briefings Bioinformat. 22(1), 30–44 (2021)
A. Bernasconi, A. Canakoglu et al., A review on viral data sources and search systems for perspective mitigation of COVID-19. Briefings in Bioinformat. 22(2), 664–675 (2021)
A. Bernasconi, A. Gulino et al., VirusViz: comparative analysis and effective visualization of viral nucleotide and aminoacid variants. Nucleic Acids Res. 49(15), e90 (2021). https://doi.org/10.1093/nar/gkab478
A. Canakoglu, A. Bernasconi et al., GenoSurf: metadata driven semantic search system for integrated genomic datasets. Database (2019)
A. Canakoglu, P. Pinoli et al., ViruSurf: an integrated database to investigate viral sequences. Nucleic Acids Res 49(D1), D817–D824 (2021)
G. Cannizzaro, M. Leone, et al., Automated integration of genomic metadata with sequence-to-sequence models. in Joint European Conference on Machine Learning and Knowledge Discovery in Databases. (Springer, Cham, 2020), pp. 187–203
E. Cappelli, F. Cumbo et al., OpenGDC: unifying, modeling, integrating cancer genomic data and Clinical Metadata. Appl. Sci. 10(18), 6367 (2020)
S. Ceri, A. Bernasconi et al., Overview of GeCo: a project for exploring and integrating signals from the genome, in Data Analytics and Management in Data Intensive Domains (DAMDID/RCDL 2017) (Springer, Cham, 2018), pp. 46–57
P. Covari, S. Pidò et al., GeCoAgent: a conversational agent for empowering genomic data extraction and analysis. in ACM Transactions on Computing for Healthcare (HEALTH) (2021)
C.A. Davis, B.C. Hitz et al., The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic acids Res. 46(D1), D794–D801 (2018)
R.L. Grossman, A.P. Heath et al., Toward a shared vision for cancer genomic data. New England J. Med. 375(12), 1109–1112 (2016)
A. Kundaje, W. Meuleman et al., Integrative analysis of 111 reference human epigenomes. Nature 518(7539), 317–330 (2015)
M. Masseroli, A. Kaitoua et al., Modeling and interoperability of heterogeneous genomic big data for integrative processing and querying. Methods 111, 3–11 (2016)
E.W. Sayers, M. Cavanaugh et al., GenBank. Nucleic Acids Res. 47(D1), D94–D99 (2019)
S.C. Schuster, Next-generation sequencing transforms today’s biology. Nature methods 5(1), 16 (2007)
Y. Shu, J. McCauley, GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance 22(13) (2017)
Z. Stark, L. Dolman et al., Integrating genomics into healthcare: a global responsibility. Am. J. Human. Genet. 104(1), 13–20 (2019)
Z.D. Stephens, S.Y. Lee et al., Big Data: Astronomical or Genomical? PLOS Biol. 13(7), 1–11 (2015)
The COVID-19 Genomics UK (COG-UK) consortium, An integrated national scale SARS-CoV-2 genomic surveillance network. The Lancet Microbe 1(3), E99–E100 (2020)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this chapter
Cite this chapter
Bernasconi, A. (2022). Model, Integrate, Search... Repeat: A Sound Approach to Building Integrated Repositories of Genomic Data. In: Piroddi, L. (eds) Special Topics in Information Technology. SpringerBriefs in Applied Sciences and Technology(). Springer, Cham. https://doi.org/10.1007/978-3-030-85918-3_8
Download citation
DOI: https://doi.org/10.1007/978-3-030-85918-3_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-85917-6
Online ISBN: 978-3-030-85918-3
eBook Packages: EngineeringEngineering (R0)