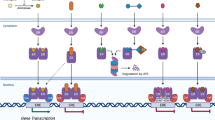
Abstract
Breast cancer is a hormone-dependent disease. Despite great strides, estrogen receptor α (ER)-positive breast cancer remains a challenging disease. About 60–70% of breast cancers are ER positive, which is predictive of response to endocrine therapies, including selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs). However, many patients will still develop therapeutic resistance. One mechanism of resistance is the development of gain-of-function mutations in the ligand-binding domain (LBD) of ER. Mutant ER exhibits ligand-independent, pro-metastatic activity, and higher concentrations of antiestrogens are required to inhibit its activity. Fulvestrant, currently the only FDA-approved SERD, possesses dose-limiting pharmacological properties and promotes only partial degradation of ER. New orally bioavailable SERMs and SERDs are being developed to overcome the shortcomings of current mainstay treatments but are challenging classes of drug to develop. Taking a ligand-binding domain-independent approach by modulating molecular chaperones and E3 ligases that control ER stability could be an alternative approach to circumvent endocrine resistance and could also be used to target additional oncogenic drivers in mutant ER tumors.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019; https://doi.org/10.3322/caac.21551.
Allen E, Doisy EA. An ovarian hormone: preliminary report on its localization, extraction and partial purification, and action in test animals. J Am Med Assoc. 1923;81(10):819–21. https://doi.org/10.1001/jama.1923.02650100027012.
Jensen EV, Jordan VC. The estrogen receptor: a model for molecular medicine. Clin Cancer Res. 2003;9(6):1980–9.
Jensen EV, Jacobson HI, Walf AA, Frye CA. Estrogen action: a historic perspective on the implications of considering alternative approaches. Physiol Behav. 2010; https://doi.org/10.1016/j.physbeh.2009.08.013.
Green S, Walter P, Greene G, et al. Cloning of the human oestrogen receptor cDNA. J Steroid Biochem. 1986; https://doi.org/10.1016/0022-4731(86)90035-X.
Walter P, Green S, Greene G, et al. Cloning of the human estrogen receptor cDNA. Proc Natl Acad Sci U S A. 1985; https://doi.org/10.1073/pnas.82.23.7889.
Beatson GT. On the treatment of inoperable cases of carcinoma of the mamma: suggestions for a new method of treatment, with illustrative cases. Trans Med Chir Soc Edinb. 1896;15:153–79. https://pubmed.ncbi.nlm.nih.gov/29584099
Tora L, White J, Brou C, et al. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989; https://doi.org/10.1016/0092-8674(89)90031-7.
Lees JA, Fawell SE, Parker MG. Identification of two transactivation domains in the mouse oestrogen receptor. Nucleic Acids Res. 1989; https://doi.org/10.1093/nar/17.14.5477.
Peng Y, Cao S, Kiselar J, et al. A metastable contact and structural disorder in the estrogen receptor transactivation domain. Structure. 2019; https://doi.org/10.1016/j.str.2018.10.026.
Ali S, Metzger D, Bornert JM, Chambon P. Modulation of transcriptional activation by ligand-dependent phosphorylation of the human oestrogen receptor A/B region. EMBO J. 1993; https://doi.org/10.1002/j.1460-2075.1993.tb05756.x.
Chen D, Riedl T, Washbrook E, et al. Activation of estrogen receptor α by S118 phosphorylation involves a ligand-dependent interaction with TFIIH and participation of CDK7. Mol Cell. 2000; https://doi.org/10.1016/S1097-2765(05)00004-3.
Arnold SF, Obourn JD, Jaffe H, Notides AC. Phosphorylation of the human estrogen receptor by mitogen-activated protein kinase and casein kinase II: consequence on DNA binding. J Steroid Biochem Mol Biol. 1995;55(2):163–72. https://doi.org/10.1016/0960-0760(95)00177-2.
Bunone G, Briand PA, Miksicek RJ, Picard D. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 1996;15(9):2174–83. https://doi.org/10.1002/j.1460-2075.1996.tb00571.x.
Kato S, Endoh H, Masuhiro Y, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270(5241):1491–4. https://doi.org/10.1126/science.270.5241.1491.
Green S, Kumar V, Theulaz I, Wahli W, Chambon P. The N-terminal DNA-binding “zinc finger” of the oestrogen and glucocorticoid receptors determines target gene specificity. EMBO J. 1988; https://doi.org/10.1002/j.1460-2075.1988.tb03168.x.
Mader S, Kumar V, De Verneuil H, Chambon P. Three amino acids of the oestrogen receptor are essential to its ability to distinguish an oestrogen from a glucocorticoid-responsive element. Nature. 1989; https://doi.org/10.1038/338271a0.
Zwart W, De Leeuw R, Rondaij M, Neefjes J, Mancini MA, Michalides R. The hinge region of the human estrogen receptor determines functional synergy between AF-1 and AF-2 in the quantitative response to estradiol and tamoxifen. J Cell Sci. 2010; https://doi.org/10.1242/jcs.061135.
Wang C, Fu M, Angeletti RH, et al. Direct acetylation of the estrogen receptor α hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem. 2001; https://doi.org/10.1074/jbc.M100800200.
Sentis S, Le Romancer M, Bianchin C, Rostan MC, Corbo L. Sumoylation of the estrogen receptor β hinge region regulates its transcriptional activity. Mol Endocrinol. 2005; https://doi.org/10.1210/me.2005-0042.
Berry NB, Fan M, Nephew KP. Estrogen receptor-α hinge-region lysines 302 and 303 regulate receptor degradation by the proteasome. Mol Endocrinol. 2008; https://doi.org/10.1210/me.2007-0449.
Shiau AK, Barstad D, Loria PM, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998; https://doi.org/10.1016/S0092-8674(00)81717-1.
Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988; https://doi.org/10.1126/science.3283939.
Devin-Leclerc J, Meng X, Delahaye F, Leclerc P, Baulieu EE, Catelli MG. Interaction and dissociation by ligands of estrogen receptor and Hsp90: the antiestrogen RU 58668 induces a protein synthesis-dependent clustering of the receptor in the cytoplasm. Mol Endocrinol. 1998; https://doi.org/10.1210/mend.12.6.0121.
Powell E, Wang Y, Shapiro DJ, Xu W. Differential requirements of Hsp90 and DNA for the formation of estrogen receptor homodimers and heterodimers. J Biol Chem. 2010; https://doi.org/10.1074/jbc.M110.104356.
Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988; https://doi.org/10.1016/0092-8674(88)90017-7.
Klein-Hitpaß L, Schorpp M, Wagner U, Ryffel GU. An estrogen-responsive element derived from the 5’flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell. 1986; https://doi.org/10.1016/0092-8674(86)90705-1.
Klinge CM, Jernigan SC, Smith SL, Tyulmenkov VV, Kulakosky PC. Estrogen response element sequence impacts the conformation and transcriptional activity of estrogen receptor α. Mol Cell Endocrinol. 2001; https://doi.org/10.1016/S0303-7207(01)00382-3.
Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997; https://doi.org/10.1038/42750.
Valley CC, Solodin NM, Powers GL, Ellison SJ, Alarid ET. Temporal variation in estrogen receptor-α protein turnover in the presence of estrogen. J Mol Endocrinol. 2008; https://doi.org/10.1677/JME-07-0067.
Fan M, Park A, Nephew KP. CHIP (carboxyl terminus of Hsc70-interacting protein) promotes basal and geldanamycin-induced degradation of estrogen receptor-α. Mol Endocrinol. 2005; https://doi.org/10.1210/me.2005-0111.
Wijayaratne AL, McDonnell DP. The human estrogen receptor-α is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J Biol Chem. 2001; https://doi.org/10.1074/jbc.M101097200.
Kushner PJ, Agard DA, Greene GL, et al. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000; https://doi.org/10.1016/S0960-0760(00)00108-4.
Safe S. Transcriptional activation of genes by 17β-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm. 2001; https://doi.org/10.1016/s0083-6729(01)62006-5.
McKay LI, Cidlowski JA. Molecular control of immune/inflammatory responses: interactions between nuclear factor-κB and steroid receptor-signaling pathways. Endocr Rev. 1999;20(4):435–59. https://doi.org/10.1210/edrv.20.4.0375.
Nettles KW, Bruning JB, Gil G, et al. NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses. Cell. 1998; https://doi.org/10.1016/j.mrfmmm.2005.02.028.
Chen D, Washbrook E, Sarwar N, et al. Phosphorylation of human estrogen receptor α at serine 118 by two distinct signal transduction pathways revealed by phosphorylation-specific antisera. Oncogene. 2002; https://doi.org/10.1038/sj.onc.1205420.
Font de Mora J, Brown M. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol Cell Biol. 2000; https://doi.org/10.1128/mcb.20.14.5041-5047.2000.
Levin ER. Plasma membrane estrogen receptors. Trends Endocrinol Metab. 2009; https://doi.org/10.1016/j.tem.2009.06.009.
Halsted WS. I. The results of operations for the cure of cancer of the breast performed at the Johns Hopkins Hospital from June, 1889, to January, 1894. Ann Surg. 1894;20(5):497–555. https://doi.org/10.1097/00000658-189407000-00075.
Boyd S. Sixty-eighth annual meeting of the british medical association. Br Med J. 1900;2(2077):1161–87. https://doi.org/10.1136/bmj.2.2077.1161.
Toft D, Gorski J. A receptor molecule for estrogens: isolation from the rat uterus and preliminary characterization. Proc Natl Acad Sci U S A. 1966; https://doi.org/10.1073/pnas.55.6.1574.
Greene GL, Gilna P, Waterfield M, Baker A, Hort Y, Shine J. Sequence and expression of human estrogen receptor complementary DNA. Science. 1986;231(4742):1150–4. https://doi.org/10.1126/science.3753802.
Jensen EV, Block GE, Smith S, Kyser K, DeSombre ER. Estrogen receptors and breast cancer response to adrenalectomy. Natl Cancer Inst Monogr. 1971;71:55.
Greene GL, Sobel NB, King WJ, Jensen EV. Immunochemical studies of estrogen receptors. J Steroid Biochem. 1984; https://doi.org/10.1016/0022-4731(84)90188-2.
King WJ, DeSombre ER, Jensen EV, Greene GL. Comparison of immunocytochemical and steroid-binding assays for estrogen receptor in human breast tumors. Cancer Res. 1985; https://doi.org/10.1007/978-1-4684-5242-6_24.
Harper MJ, Walpole AL. Mode of action of I.C.I. 46,474 in preventing implantation in rats. J Endocrinol. 1967; https://doi.org/10.1677/joe.0.0370083.
Skidmore J, Walpole AL, Woodburn J. Effect of some triphenylethylenes on oestradiol binding in vitro to macromolecules from uterus and anterior pituitary. J Endocrinol. 1972; https://doi.org/10.1677/joe.0.0520289.
Jordan VC. Prolonged antioestrogenic activity of ICI 46,474 in the ovariectomized mouse. J Reprod Fertil. 1975; https://doi.org/10.1530/jrf.0.0420251.
Jordan VC, Koerner S. Tamoxifen (ICI 46,474) and the human carcinoma 8S oestrogen receptor. Eur J Cancer. 1975; https://doi.org/10.1016/0014-2964(75)90119-X.
Jordan VC. Effect of tamoxifen (ICI 46,474) on initiation and growth of DMBA-induced rat mammary carcinomata. Eur J Cancer. 1976; https://doi.org/10.1016/0014-2964(76)90030-X.
Sakamoto T, Eguchi H, Omoto Y, Ayabe T, Mori H, Hayashi S. Estrogen receptor-mediated effects of tamoxifen on human endometrial cancer cells. Mol Cell Endocrinol. 2002;192(1):93–104. https://doi.org/10.1016/S0303-7207(02)00086-2.
McInerney EM, Katzenellenbogen BS. Different regions in activation function-1 of the human estrogen receptor required for antiestrogen- and estradiol-dependent transcription activation. J Biol Chem. 1996; https://doi.org/10.1074/jbc.271.39.24172.
Liu H, Lee ES, De Los Reyes A, Zapf JW, Jordan VC. Silencing and reactivation of the selective estrogen receptor modulator-estrogen receptor α complex. Cancer Res. 2001;61(9):3632–9.
Long X, Nephew KP. Fulvestrant (ICI 182,780)-dependent interacting proteins mediate immobilization and degradation of estrogen receptor-α. J Biol Chem. 2006; https://doi.org/10.1074/jbc.M510809200.
Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov. 2016; https://doi.org/10.1158/2159-8290.CD-15-0894.
Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015; https://doi.org/10.1038/nrclinonc.2015.117.
Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009; https://doi.org/10.1038/nrc2713.
Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011; https://doi.org/10.1146/annurev-med-070909-182917.
Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, et al. D538G mutation in estrogen receptor-α: a novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013; https://doi.org/10.1158/0008-5472.CAN-13-1197.
Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013; https://doi.org/10.1038/ng.2822.
Robinson DR, Wu YM, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013; https://doi.org/10.1038/ng.2823.
Jeselsohn R, Yelensky R, Buchwalter G, et al. Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor–positive breast cancer. Clin Cancer Res. 2014;20(7):1757–67. https://doi.org/10.1158/1078-0432.CCR-13-2332.
Li S, Shen D, Shao J, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013; https://doi.org/10.1016/j.celrep.2013.08.022.
Dustin D, Gu G, Fuqua SAW. ESR1 mutations in breast cancer. Cancer. 2019; https://doi.org/10.1002/cncr.32345.
Jeselsohn R, Bergholz JS, Pun M, et al. Allele-specific chromatin recruitment and therapeutic vulnerabilities of ESR1 activating mutations. Cancer Cell. 2018; https://doi.org/10.1016/j.ccell.2018.01.004.
Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol. 2016; https://doi.org/10.1001/jamaoncol.2016.1279.
Spoerke JM, Gendreau S, Walter K, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016; https://doi.org/10.1038/ncomms11579.
Fanning SW, Mayne CG, Dharmarajan V, et al. Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. elife. 2016; https://doi.org/10.7554/eLife.12792.
Zhao Y, Laws MJ, Guillen VS, et al. Structurally novel antiestrogens elicit differential responses from constitutively active mutant estrogen receptors in breast cancer cells and tumors. Cancer Res. 2017; https://doi.org/10.1158/0008-5472.CAN-17-1265.
Katzenellenbogen JA, Mayne CG, Katzenellenbogen BS, Greene GL, Chandarlapaty S. Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat Rev Cancer. 2018;18(6):377–88. https://doi.org/10.1038/s41568-018-0001-z.
Toy W, Weir H, Razavi P, et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 2017; https://doi.org/10.1158/2159-8290.CD-15-1523.
Fuqua SAW, Wiltschke C, Zhang QX, et al. A hypersensitive estrogen receptor-α mutation in premalignant breast lesions. Cancer Res. 2000;60(15):4026–9. http://cancerres.aacrjournals.org/content/60/15/4026.abstract.
Brzozowski AM, Pike ACW, Dauter Z, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997; https://doi.org/10.1038/39645.
Gates LA, Gu G, Chen Y, et al. Proteomic profiling identifies key coactivators utilized by mutant ERα proteins as potential new therapeutic targets. Oncogene. 2018; https://doi.org/10.1038/s41388-018-0284-2.
Fu X, Pereira R, De Angelis C, et al. FOXA1 upregulation promotes enhancer and transcriptional reprogramming in endocrine-resistant breast cancer. Proc Natl Acad Sci U S A. 2019; https://doi.org/10.1073/pnas.1911584116.
Harrod A, Fulton J, Nguyen VTM, et al. Genomic modelling of the ESR1 Y537S mutation for evaluating function and new therapeutic approaches for metastatic breast cancer. Oncogene. 2017; https://doi.org/10.1038/onc.2016.382.
Martin LA, Ribas R, Simigdala N, et al. Discovery of naturally occurring ESR1 mutations in breast cancer cell lines modelling endocrine resistance. Nat Commun. 2017; https://doi.org/10.1038/s41467-017-01864-y.
Andreano KJ, Baker JG, Park S, et al. The dysregulated pharmacology of clinically relevant ESR1 mutants is normalized by ligand-activated WT receptor. Mol Cancer Ther. 2020; https://doi.org/10.1158/1535-7163.mct-19-1148.
Helzer KT, Szatkowski Ozers M, Meyer MB, et al. The phosphorylated estrogen receptor α (ER) cistrome identifies a subset of active enhancers enriched for direct ER-DNA binding and the transcription factor GRHL2. Mol Cell Biol. 2018; https://doi.org/10.1128/mcb.00417-18.
Rajbhandari P, Schalper KA, Solodin NM, et al. Pin1 modulates ERα levels in breast cancer through inhibition of phosphorylation-dependent ubiquitination and degradation. Oncogene. 2014; https://doi.org/10.1038/onc.2013.78.
Jia S, Miedel MT, Ngo M, et al. Clinically observed estrogen receptor alpha mutations within the ligand-binding domain confer distinguishable phenotypes. Oncologia. 2018; https://doi.org/10.1159/000485510.
Sun J, Zhou W, Kaliappan K, Nawaz Z, Slingerland JM. ERα phosphorylation at Y537 by Src triggers E6-AP-ERα binding, ERα ubiquitylation, promoter occupancy, and target gene expression. Mol Endocrinol. 2012; https://doi.org/10.1210/me.2012-1140.
Fuqua SAW, Gu G, Rechoum Y, et al. Abstract S4-02: The Y537S ESR1 mutation is a dominant driver of distant ER-positive breast cancer metastasis. Cancer Res. 2017;77(4 Supplement):S4-02. https://doi.org/10.1158/1538-7445.SABCS16-S4-02.
O’leary B, Cutts RJ, Liu Y, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 2018; https://doi.org/10.1158/2159-8290.CD-18-0264.
Schiavon G, Hrebien S, Garcia-Murillas I, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015; https://doi.org/10.1126/scitranslmed.aac7551.
Zhang K, Hong R, Xu F, et al. Clinical value of circulating ESR1 mutations for patients with metastatic breast cancer: a meta-analysis. Cancer Manag Res. 2018; https://doi.org/10.2147/CMAR.S173193.
Najim O, Seghers S, Sergoynne L, et al. The association between type of endocrine therapy and development of estrogen receptor-1 mutation(s) in patients with hormone-sensitive advanced breast cancer: a systematic review and meta-analysis of randomized and non-randomized trials. Biochim Biophys Acta. 2019; https://doi.org/10.1016/j.bbcan.2019.188315.
De Santo I, McCartney A, Malorni L, Migliaccio I, Di Leo A. The emerging role of esr1 mutations in luminal breast cancer as a prognostic and predictive biomarker of response to endocrine therapy. Cancers (Basel). 2019; https://doi.org/10.3390/cancers11121894.
Linden HM, Peterson LM, Fowler AM. Clinical potential of estrogen and progesterone receptor imaging. PET Clin. 2018;13(3):415–22. https://doi.org/10.1016/j.cpet.2018.02.005.
Kumar M, Salem K, Michel C, Jeffery JJ, Yan Y, Fowler AM. 18F-fluoroestradiol PET imaging of activating estrogen receptor-a mutations in breast cancer. J Nucl Med. 2019; https://doi.org/10.2967/jnumed.118.224667.
Laine M, Greene M, Chang Y-F, et al. Abstract PD7-09: Lasofoxifene decreases breast cancer lung and liver metastasis in a mammary intraductal (MIND) xenograft model of mutant ERα+ breast cancer. Cancer Res. 2019; https://doi.org/10.1158/1538-7445.sabcs18-pd7-09.
Fanning SW, Jeselsohn R, Dharmarajan V, et al. The SERM/SERD bazedoxifene disrupts ESR1 helix 12 to overcome acquired hormone resistance in breast cancer cells. elife. 2018; https://doi.org/10.7554/eLife.37161.
Wardell SE, Nelson ER, Chao CA, McDonnell DP. Bazedoxifene exhibits antiestrogenic activity in animal models of tamoxifen-resistant breast cancer: implications for treatment of advanced disease. Clin Cancer Res. 2013; https://doi.org/10.1158/1078-0432.CCR-12-3771.
Wardell SE, Ellis MJ, Alley HM, et al. Efficacy of SERD/SERM hybrid-CDK4/6 inhibitor combinations in models of endocrine therapy-resistant breast cancer. Clin Cancer Res. 2015; https://doi.org/10.1158/1078-0432.CCR-15-0360.
Nardone A, Weir H, Delpuech O, et al. The oral selective oestrogen receptor degrader (SERD) AZD9496 is comparable to fulvestrant in antagonising ER and circumventing endocrine resistance. Br J Cancer. 2019; https://doi.org/10.1038/s41416-018-0354-9.
Weir HM, Bradbury RH, Rabow AA, et al. AZD9496: an oral estrogen receptor inhibitor that blocks the growth of ER-positive and ESR1-mutant breast tumors in preclinical models. Cancer Res. 2016; https://doi.org/10.1158/0008-5472.CAN-15-2357.
Hamilton EP, Oliveira M, Banerji U, et al. A phase I dose escalation and expansion study of the next generation oral SERD AZD9833 in women with ER-positive, HER2-negative advanced breast cancer. J Clin Oncol. 2020; https://doi.org/10.1200/jco.2020.38.15_suppl.1024.
Lu Y, Gutgesell LM, Xiong R, et al. Design and synthesis of basic selective estrogen receptor degraders for endocrine therapy resistant breast cancer. J Med Chem. 2019; https://doi.org/10.1021/acs.jmedchem.9b01580.
Zhang L, Dai D, Shi Z, Jiang J, Wang Y. Abstract OT1-01-04: Phase 1 study of D-0502, an orally bioavailable SERD with optimized pharmacological and PK/PD property for ER-positive breast cancer. Cancer Res. 2019;79(4 Supplement):OT1-01-04. https://doi.org/10.1158/1538-7445.SABCS18-OT1-01-04.
Andreano KJ, Wardell SE, Baker JG, et al. G1T48, an oral selective estrogen receptor degrader, and the CDK4/6 inhibitor lerociclib inhibit tumor growth in animal models of endocrine-resistant breast cancer. Breast Cancer Res Treat. 2020; https://doi.org/10.1007/s10549-020-05575-9.
Metcalfe C, Ingalla E, Blake R, et al. Abstract P5-04-07: GDC-9545: a novel ER antagonist and clinical candidate that combines desirable mechanistic and pre-clinical DMPK attributes. Cancer Res. 2019; https://doi.org/10.1158/1538-7445.sabcs18-p5-04-07.
Guan J, Zhou W, Hafner M, et al. Therapeutic ligands antagonize estrogen receptor function by impairing its mobility. Cell. 2019; https://doi.org/10.1016/j.cell.2019.06.026.
Joseph JD, Darimont B, Zhou W, et al. The selective estrogen receptor downregulator GDC-0810 is efficacious in diverse models of ER+ breast cancer. elife. 2016; https://doi.org/10.7554/eLife.15828.
Lai A, Kahraman M, Govek S, et al. Identification of GDC-0810 (ARN-810), an orally bioavailable selective estrogen receptor degrader (SERD) that demonstrates robust activity in tamoxifen-resistant breast cancer xenografts. J Med Chem. 2015; https://doi.org/10.1021/acs.jmedchem.5b00054.
Tria GS, Abrams T, Baird J, et al. Discovery of LSZ102, a potent, orally bioavailable selective estrogen receptor degrader (SERD) for the treatment of estrogen receptor positive breast cancer. J Med Chem. 2018; https://doi.org/10.1021/acs.jmedchem.7b01682.
Bihani T, Patel HK, Arlt H, et al. Elacestrant (RAD1901), a Selective Estrogen Receptor Degrader (SERD), has antitumor activity in multiple ER+ breast cancer patient-derived xenograft models. Clin Cancer Res. 2017; https://doi.org/10.1158/1078-0432.CCR-16-2561.
Patel HK, Tao N, Lee KM, et al. Elacestrant (RAD1901) exhibits anti-tumor activity in multiple ER+ breast cancer models resistant to CDK4/6 inhibitors. Breast Cancer Res. 2019; https://doi.org/10.1186/s13058-019-1230-0.
Garner F, Shomali M, Paquin D, Lyttle CR, Hattersley G. RAD1901: a novel, orally bioavailable selective estrogen receptor degrader that demonstrates antitumor activity in breast cancer xenograft models. Anti-Cancer Drugs. 2015; https://doi.org/10.1097/CAD.0000000000000271.
Wardell SE, Nelson ER, Chao CA, Alley HM, McDonnell DP. Evaluation of the pharmacological activities of RAD1901, a selective estrogen receptor degrader. Endocr Relat Cancer. 2015; https://doi.org/10.1530/ERC-15-0287.
Campone M, Bardia A, Ulaner GA, et al. Phase I/II study of SAR439859, an oral selective estrogen receptor degrader (SERD), in estrogen receptor-positive (ER+)/human epidermal growth factor receptor 2-negative (HER2-) metastatic breast cancer (mBC). J Clin Oncol. 2020; https://doi.org/10.1200/jco.2020.38.15_suppl.1070.
Shomali M, Cheng J, Koundinya M, et al. Abstract P3-04-05: identification of SAR439859, an orally bioavailable selective estrogen receptor degrader (SERD) that has strong antitumor activity in wild-type and mutant ER+ breast cancer models. Cancer Res. 2017; https://doi.org/10.1158/1538-7445.sabcs16-p3-04-05.
Bardia A, Linden HM, Ulaner GA, Chandarlapaty S, Gosselin A, Celanovic M, Campone M. Phase 1/2 dose-escalation and expansion study investigating SAR439859 +/-palbociclib in postmenopausal women with estrogen receptor-positive (ER+)/HER2-metastatic breast cancer. J Clin Oncol. 2019; https://doi.org/10.1200/JCO.2019.37.15-suppl.TPS1105.
Puyang X, Furman C, Zheng GZ, et al. Discovery of selective estrogen receptor covalent antagonists for the treatment of ERαWT and ERαMUT breast cancer. Cancer Discov. 2018; https://doi.org/10.1158/2159-8290.CD-17-1229.
Hamilton EP, Dees EC, Wang JS-Z, et al. Phase I dose escalation of H3B-6545, a first-in-class highly Selective ERα Covalent Antagonist (SERCA), in women with ER-positive, HER2-negative breast cancer (HR+ BC). J Clin Oncol. 2019;37(15_suppl):1059. https://doi.org/10.1200/JCO.2019.37.15_suppl.1059.
Rioux N, Smith S, Korpal M, et al. Nonclinical pharmacokinetics and in vitro metabolism of H3B-6545, a novel selective ERα covalent antagonist (SERCA). Cancer Chemother Pharmacol. 2019; https://doi.org/10.1007/s00280-018-3716-3.
Roberts BL, Ma Z-X, Gao A, et al. Two-stage strategy for development of proteolysis targeting chimeras and its application for estrogen receptor degraders. ACS Chem Biol. 2020; https://doi.org/10.1021/acschembio.0c00140.
Flanagan J, Qian Y, Gough S, et al. Abstract P5-04-18: ARV-471, an oral estrogen receptor PROTAC degrader for breast cancer. Cancer Res. 2019; https://doi.org/10.1158/1538-7445.sabcs18-p5-04-18.
Gonzalez TL, Hancock M, Sun S, et al. Targeted degradation of activating estrogen receptor α ligand-binding domain mutations in human breast cancer. Breast Cancer Res Treat. 2020; https://doi.org/10.1007/s10549-020-05564-y.
Zhao Z, Wang L, James T, et al. Reciprocal regulation of ERα and ERβ stability and activity by diptoindonesin G. Chem Biol. 2015; https://doi.org/10.1016/j.chembiol.2015.10.011.
Liu JT, Do TJ, Simmons CJ, et al. Total synthesis of diptoindonesin G and its analogues as selective modulators of estrogen receptors. Org Biomol Chem. 2016; https://doi.org/10.1039/c6ob01657j.
Gao J, Fan M, Xiang G, et al. Diptoindonesin G promotes ERK-mediated nuclear translocation of p-STAT1 (Ser727) and cell differentiation in AML cells. Cell Death Dis. 2017; https://doi.org/10.1038/cddis.2017.159.
Fan M, Chen J, Gao J, et al. Triggering a switch from basal- to luminal-like breast cancer subtype by the small-molecule diptoindonesin G via induction of GABARAPL1. Cell Death Dis. 2020; https://doi.org/10.1038/s41419-020-02878-z.
Wardell SE, Yllanes AP, Chao CA, et al. Pharmacokinetic and pharmacodynamic analysis of fulvestrant in preclinical models of breast cancer to assess the importance of its estrogen receptor-α degrader activity in antitumor efficacy. Breast Cancer Res Treat. 2020; https://doi.org/10.1007/s10549-019-05454-y.
van Kruchten M, de Vries EG, Glaudemans AW, et al. Measuring residual estrogen receptor availability during fulvestrant therapy in patients with metastatic breast cancer. Cancer Discov. 2015; https://doi.org/10.1158/2159-8290.CD-14-0697.
Robertson JFR. Fulvestrant (Faslodex®)—how to make a good drug better. Oncologist. 2007; https://doi.org/10.1634/theoncologist.12-7-774.
Robertson JFR, Harrison M. Fulvestrant: pharmacokinetics and pharmacology. Br J Cancer. 2004; https://doi.org/10.1038/sj.bjc.6601630.
Tikoo D, Gupta M. Duavee: a tissue-selective estrogen complex for menopausal symptoms and prevention of osteoporosis. Int J Basic Clin Pharmacol. 2015; https://doi.org/10.5455/2319-2003.ijbcp20150425.
Lewis-Wambi JS, Kim H, Curpan R, Grigg R, Sarker MA, Jordan VC. The selective estrogen receptor modulator bazedoxifene inhibits hormone-independent breast cancer cell growth and down-regulates estrogen receptor α and cyclin D1. Mol Pharmacol. 2011; https://doi.org/10.1124/mol.111.072249.
Komm BS, Kharode YP, Bodine PVN, Harris HA, Miller CP, Lyttle CR. Bazedoxifene acetate: a selective estrogen receptor modulator with improved selectivity. Endocrinology. 2005; https://doi.org/10.1210/en.2005-0030.
Bardia A, Aftimos P, Bihani T, et al. EMERALD: phase III trial of elacestrant (RAD1901) vs endocrine therapy for previously treated ER+ advanced breast cancer. Future Oncol. 2019; https://doi.org/10.2217/fon-2019-0370.
Kaklamani V, Bardia A, Wilks S, et al. Abstract PD7-07: final analysis of phase 1 study of elacestrant (RAD1901), a novel selective estrogen receptor degrader (SERD), in estrogen receptor positive (ER+), human epidermal growth factor receptor 2 negative (HER2-) advanced breast cancer. J Clin Oncol. 2020; https://doi.org/10.1158/1538-7445.sabcs19-pd7-07.
Sreekumar S, Levine K, Sikora M, et al. Differential regulation and targeting of estrogen receptor α turnover in invasive lobular breast carcinoma. Endocrinology. 2020; https://doi.org/10.1210/endocr/bqaa109.
Wardell SE, Marks JR, McDonnell DP. The turnover of estrogen receptor α by the selective estrogen receptor degrader (SERD) fulvestrant is a saturable process that is not required for antagonist efficacy. Biochem Pharmacol. 2011; https://doi.org/10.1016/j.bcp.2011.03.031.
Bosch A, Li Z, Bergamaschi A, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med. 2015; https://doi.org/10.1126/scitranslmed.aaa4442.
Fribbens C, O’Leary B, Kilburn L, et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2016; https://doi.org/10.1200/JCO.2016.67.3061.
Gelsomino L, Gu G, Rechoum Y, et al. ESR1 mutations affect anti-proliferative responses to tamoxifen through enhanced cross-talk with IGF signaling. Breast Cancer Res Treat. 2016; https://doi.org/10.1007/s10549-016-3829-5.
Li Z, Levine KM, Bahreini A, et al. Upregulation of IRS1 enhances IGF1 response in Y537S and D538G ESR1 mutant breast cancer cells. Endocrinology. 2018; https://doi.org/10.1210/en.2017-00693.
Nayar U, Cohen O, Kapstad C, et al. Acquired HER2 mutations in ER + metastatic breast cancer confer resistance to estrogen receptor–directed therapies. Nat Genet. 2019; https://doi.org/10.1038/s41588-018-0287-5.
Mao P, Cohen O, Kowalski KJ, et al. Acquired FGFR and FGF alterations confer resistance to estrogen receptor (ER) targeted therapy in ER+ metastatic breast cancer. bioRxiv. 2019:605436. https://doi.org/10.1101/605436.
Tateishi Y, Kawabe YI, Chiba T, et al. Ligand-dependent switching of ubiquitin-proteasome pathways for estrogen receptor. EMBO J. 2004; https://doi.org/10.1038/sj.emboj.7600472.
Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010; https://doi.org/10.1038/nrc2887.
Lai BT, Chin NW, Stanek AE, Keh W, Lanks KW. Quantitation and intracellular localization of the 85K heat shock protein by using monoclonal and polyclonal antibodies. Mol Cell Biol. 1984;4(12):2802–10. https://doi.org/10.1128/MCB.4.12.2802.
Grenert JP, Johnson BD, Toft DO. The importance of ATP binding and hydrolysis by Hsp90 in formation and function of protein heterocomplexes. J Biol Chem. 1999;274(25):17525–33. https://doi.org/10.1074/jbc.274.25.17525.
Catelli MG, Binart N, Jung-Testas I, et al. The common 90-kd protein component of non-transformed “8S” steroid receptors is a heat-shock protein. EMBO J. 1985;4(12):3131–5. https://pubmed.ncbi.nlm.nih.gov/2419124.
Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and Immunophilin chaperones*. Endocr Rev. 1997;18(3):306–60. https://doi.org/10.1210/edrv.18.3.0303.
Kamal A, Thao L, Sensintaffar J, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003; https://doi.org/10.1038/nature01913.
Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90(1):65–75. https://doi.org/10.1016/S0092-8674(00)80314-1.
Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89(2):239–50. https://doi.org/10.1016/S0092-8674(00)80203-2.
Obermann WMJ, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143(4):901–10. https://doi.org/10.1083/jcb.143.4.901.
Panaretou B, Prodromou C, Roe SM, et al. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17(16):4829–36. https://doi.org/10.1093/emboj/17.16.4829.
Patricia Hernández M, Sullivan WP, Toft DO. The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J Biol Chem. 2002; https://doi.org/10.1074/jbc.M206566200.
Bagatell R, Khan O, Paine-Murrieta G, Taylor CW, Akinaga S, Whitesell L. Destabilization of steroid receptors by heat shock protein 90-binding drugs: a ligand-independent approach to hormonal therapy of breast cancer. Clin Cancer Res. 2001;7(7):2076–84.
Zhou W, Slingerland JM. Links between oestrogen receptor activation and proteolysis: relevance to hormone-regulated cancer therapy. Nat Rev Cancer. 2014; https://doi.org/10.1038/nrc3622.
Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. 2020; https://doi.org/10.1038/s41392-020-0107-0.
Saji S, Okumura N, Eguchi H, et al. MDM2 enhances the function of estrogen receptor in human breast cancer cells. Biochem Biophys Res Commun. 2001; https://doi.org/10.1006/bbrc.2001.4339.
Hashizume R, Fukuda M, Maeda I, et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001; https://doi.org/10.1074/jbc.C000881200.
Eakin CM, MacCoss MJ, Finney GL, Klevit RE. Estrogen receptor α is a putative substrate for the BRCA1 ubiquitin ligase. Proc Natl Acad Sci U S A. 2007; https://doi.org/10.1073/pnas.0610887104.
Bhatt S, Xiao Z, Meng Z, Katzenellenbogen BS. Phosphorylation by p38 mitogen-activated protein kinase promotes estrogen receptor turnover and functional activity via the SCFSkp2 proteasomal complex. Mol Cell Biol. 2012; https://doi.org/10.1128/mcb.06561-11.
Yu M, Bardia A, Aceto N, et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014; https://doi.org/10.1126/science.1253533.
Ito T, Ando H, Suzuki T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010; https://doi.org/10.1126/science.1177319.
Singhal S, Mehta J, Desikan R, et al. Antitumor activity of thalidomide in refractory multiple myeloma. N Engl J Med. 1999; https://doi.org/10.1056/NEJM199911183412102.
Gandhi AK, Kang J, Havens CG, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4CRBN. Br J Haematol. 2014; https://doi.org/10.1111/bjh.12708.
Hansen JD, Correa M, Nagy MA, et al. Discovery of CRBN E3 ligase modulator CC-92480 for the treatment of relapsed and refractory multiple myeloma. J Med Chem. 2020;63(13):6648–76. https://doi.org/10.1021/acs.jmedchem.9b01928.
Funding
This work was supported by NIH R01s CA213292 and CA236356, DOD BC190650 to W.X. and NIH T32 CA009135 to K. D.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Donahue, K., Xu, W. (2021). Therapeutic Strategies to Target Activating Estrogen Receptor α Mutations. In: Badr, M.Z. (eds) Nuclear Receptors. Springer, Cham. https://doi.org/10.1007/978-3-030-78315-0_15
Download citation
DOI: https://doi.org/10.1007/978-3-030-78315-0_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-78314-3
Online ISBN: 978-3-030-78315-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)