Abstract
Bayesian network modelling is applied to health psychology data in order to obtain more insight into the determinants of physical activity. This preliminary study discusses some challenges to apply general machine learning methods to this application domain, and Bayesian networks in particular. We investigate several suitable methods for dealing with missing data, and determine which method obtains good results in terms of fitting the data. Furthermore, we present the learnt Bayesian network model for this e-health intervention case study, and conclusions are drawn about determinants of physical activity behaviour change and how the intervention affects physical activity behaviour and its determinants. We also evaluate the contributions of Bayesian network analysis compared to traditional statistical analyses in this field. Finally, possible extensions on the performed analyses are proposed.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Nowadays there are various e-health intervention platforms that employ integrated behaviour change techniques in order to change health-related-behaviour of participants, for example increasing physical activity. These interventions apply theoretical psychological methods to influence behavioural determinants, which are factors determining a certain behaviour. These general techniques are translated to behaviour change strategies by tailoring the theoretical method to the target population and intervention setting [1]. To measure the effects of such interventions, various research studies have been performed, assessing physical activity with tools such as questionnaires and activity trackers. While there is now a good understanding of what the most important determinants for increasing physical activity are, little is known about how these determinants interact. Improved understanding of these relationships could be used to improve existing e-health interventions.
Supervised machine learning techniques are used to identify relationships underlying data with labeled input and output, and predict output results for a given input. These techniques could for example be used to model relations between diseases and symptoms and give expectations about the presence of various diseases given symptoms. Bayesian networks [8] represent probabilistic relationships between a set of variables, where relationships between the input variables can also be investigated. Such networks can make probabilistic predictions and provide a visual insight in relations among all variables of interest, thereby providing a potential useful tool to exploratively investigate and better understand determinants of physical activity.
In this article, a Bayesian network model is learned from data from a single intervention study, i.e., the Active Plus intervention [12], aiming at influencing physical activity behaviour among older adults. We discuss ways to learn from these complex data containing a significant amount of missing values. Based on these initial findings, results from previous analyses are compared to results from applying the Bayesian network model to the same data, to examine the added value of this technique compared to traditional ones. We show that learning a Bayesian network model for measurement data from the Active Plus project indeed reveals conditional dependence and independence relations that provide new insights and explanations for previously found results.
This paper is organised as follows. Section 2 provides technical background about methods and algorithms. Section 3 provides a description of the data and intervention study at hand, and how the data has been pre-processed. Furthermore, the analysis based on the Bayesian network model is explained including a description of the applied learning strategy, and a missing data analysis to select appropriate methods for handling the missing data. Then, in Sect. 4, results are given about the comparison of evaluated methods, and the comparison of the results from the Bayesian network model, determined using the best method, and the results from previous analyses. Finally, Sect. 5 concludes this paper and Sect. 6 elaborates on possible extensions.
2 Preliminaries
This section gives an overview of the theoretical background relevant to perform the case study analyses, including a brief introduction of the modelling approach.
2.1 Bayesian Network Model
A Bayesian network [8] is a probabilistic graphical model represented as a directed acyclic graph \(G=(V,E)\), where the set of nodes V represent random variables, and the set of arcs E represent probabilistic independencies among the variables. Associated with each node is a conditional probability distribution of that variable given its parents. The graphical structure implies conditional independence statements. Let \(V = \{X_1, \ldots , X_n\}\) be an enumeration of the nodes in a Bayesian network such that each node appears after its children, and let \(\varPi _{i}\) be the set of parents of a node \(X_i\). The local Markov property in the Bayesian network states that \(X_i\) is conditionally independent of all variables in \(\{ X_1, X_2, \ldots , X_{i-1} \}\) given \(\varPi _{i}\) for all \(i \in \{1, \ldots , n\}\). These local independences imply conditional independence statements over arbitrary sets of variables.
The joint probability distribution over discrete variables follows from the conditional independence propositions and conditional probabilities:
where the first equation follows from the usual chain rule in probability theory and the second from the local Markov property. Note that the conditional probabilities \(\mathbb {P}(X_i\mid \varPi _{i})\) correspond to the arcs in the Bayesian network specification. In continuous Bayesian networks, usually a linear Gaussian distribution is assumed, where the joint density is factorised where each \(X_i \mid \varPi _i \sim \mathcal {N}(\beta \varPi _i + \alpha , \sigma ^2)\).
A temporal Bayesian network is an extension to the static counterpart in that it is a Bayesian network model over time, where the nodes represent the random variables occurring at particular time slices. The temporal Bayesian network model is subject to the condition that arcs directed to variables in previous time slices cannot occur. In case the temporal Bayesian network is time-homogeneous (or time-invariant), these models are also called dynamic Bayesian networks [6]. Since in this case study there are only a few time slices and differences between these slices are not constant, we do not assume time-invariance in the remainder of this paper.
2.2 Learning Bayesian Networks
The following three common classes of algorithms are used to learn the structure of Bayesian networks from the data: constraint-based algorithms which employ conditional independence tests to learn the dependence structure of the data, score-based algorithms which use search algorithms to find a graph that maximises a goodness-of-fit scores as objective function, and hybrid algorithms which combine both approaches. Recent research has shown that constraint-based algorithms are often less accurate and seldom faster and hybrid algorithms are neither faster nor more accurate [11]. For this reason, we focus in the remainder of this paper on score-based structure learning algorithms, where local search methods are used to explore the space of directed acyclic graphs by single-arc addition, removal and reversal. In particular, we apply tabu search to the physical activity data in this case study as empirical evidence shows that this search method typically performs well for learning Bayesian networks [5, Chapter 13.7].
There are several model selection criteria that are used in the search-based structure learning algorithms , where in this paper we have chosen the commonly-used Bayesian Information Criterion (BIC) [9]. To fit the parameters we have chosen a uniform prior distribution over the model parameters [4].

2.3 Handling Missing Data
Learning Bayesian networks with missing data is significantly harder as the log-likelihood does not admit a closed-form solution if values are missing. In this paper, we assume that data are missing at random, for which commonly used methods are listwise deletion, pair-wise deletion, single imputation, multiple imputation [7]. The deletion approaches omit (observed) values from analyses. In the listwise deletion approach on the one hand, all observations with missing values at any measurement are omitted completely. On the other hand, the pair-wise deletion method does not require complete data on all variables in the model, and mean and covariance estimations are here based on the full number of observations with complete data for each (pair of) variable(s). Imputation methods involve replacing missing values by estimates such as by the mean of observed values in the attribute, called mean imputation. Single imputation imputes a single value treating it as known, whereas multiple imputation replaces missing values by two or more values representing a distribution of possibilities. In multiple imputation, missing data are filled in an arbitrary number of times to generate different complete datasets to be analysed, and results are combined for inference. Finally, in Bayesian network learning, the Expectation Maximization (EM) algorithm [2] is often applied, which iteratively optimises parameters in order to find the maximum likelihood estimate, assuming the missing data is missing at random (MAR). The Structural EM algorithm (SEM) [3] combines this standard EM algorithm with structure search for model selection.
The variant of the structural EM algorithm that is used in this case study can be described as follows (see Algorithm 1 for an overview). Let \(\mathbf {d}\) be a dataset over the set of random variables \(\mathbf {V}\). Assume that \(\mathbf {o}\) is part of the dataset that is actually observed, i.e., \(\mathbf {o} \subseteq \mathbf {d}\). Furthermore, we denote the missing data by \(\mathbf {h}\), i.e., \(\mathbf {d} = \mathbf {o} \cup \mathbf {h}\), and \(\mathbf {o} \cap \mathbf {h} = \varnothing \). The SEM algorithm aims to find a model from the space of Bayesian network models over \(\mathbf {V}\), denoted by \(\mathcal {M}\), such that each model \(M\in \mathcal {M}\) is parametrised by a vector \(\varTheta ^M\) defining a probability distribution \(\mathbb {P}(\mathbf {V}:M,\varTheta ^M)\). To find a model in case of missing values, the complete data likelihood \(\mathbb {P}(\mathbf {H}, \mathbf {O} \mid M )\) is estimated. The algorithm iteratively maximises the expected Bayesian network model score optimised by the score-based algorithm. First the posterior parameter distributions, given the currently best model structure and observed data, are computed. In the expectation step, these distributions are used to compute the expected complete dataset, imputing missing values with their most probable values, also sometimes called hard EM. During the maximization, the currently best model structure is updated using a tabu structure learning algorithm, using the imputed data from the expectation step. Then parameter learning gives new distributions to be used as input for the next expectation step. To perform the first expectation, an initial network structure is given as input to the algorithm. In case a maximum number of iterations is reached or in case of convergence, the Bayesian network model is returned.
3 Description of the Data and Methodology
The experiments in this intervention case study aim to analyse performance of different methods to handle missing values and to learn the Bayesian network model for given intervention data in order to compare its results to previous analyses. This section describes the data, preprocessing phase, magnitude of the missing data problem and the approach to determine a suitable method in order to analyse the data by Bayesian network learning. The raw research data that has been collected during the Active Plus intervention was provided to the authors and is described in the first subsection.
3.1 Data Acquisition and Description
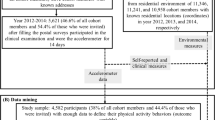
The raw research data has mostly been collected via questionnaires and consists of determinants, external factors, measurements of physical activity and intervention-related information at different time-slots, starting with a baseline measurement before the participant receives the intervention [14]. For example, the validated self-administrated Dutch Short Questionnaire to Assess Health Enhancing Physical Activity (SQUASH) is included in the questionnaires as subjective measurement of physical activity [16]. Figure 1 illustrates the intervention outline including moments of receiving intervention content and of measurement in time [12]. There is a distinction between control, intervention basic and intervention-plus groups, representing the intervention condition. This condition determines whether a participant receives an intervention or not and if environmental content is included in the intervention with additional information such as opportunities to be physically active in the own environment. Within these main groups, content is further personalised based on characteristics of participants, for example state of behaviour change (stage) measured at baseline or age. Since in the analyses in this article intervention content is proxied by a few main characteristics, this personalisation is beyond the focus of this article [12].
Outline intervention program including moments of measurement [12].
As depicted in Fig. 1, data has been collected at 4 time-slots; at the baseline (before receiving the intervention, T0) and, to measure intervention effects, 3 (T1), 6 (T2) and 12 (T3) months after the baseline. About 1258 variables have been measured for a sub-population being a random sample of 1976 adults aged 50 and older. Measurements are at item-level of detail, where an item is a specific measurement, for example a question in the questionnaire. In preprocessing rules, it is described how concepts are calculated from item data in order to perform analyses at a higher level of abstraction.
3.2 Data Preprocessing and Concept Design
The raw data is preprocessed, according to rules to integrate data from different studies and to aggregate, by calculating concepts from the raw data at item-level of detail, as mentioned in the previous subsection. This subsection describes assumptions and decisions made during the data preprocessing phase and rules to calculate the concepts included in analyses in this article.
In general, concepts are calculated by the mean or sum of items taking into account a maximum percentage of items allowed to be missing, except from a few concepts calculated using predefined formulas. In particular, the SQUASH-outcome measure, which is the number of minutes per week of moderate to intensive physical activity, is calculated in a standardised way [12]. In case more than 25% of the items are missing, the concept value is assumed to be missing. Besides these aggregation rules, preprocessing rules contain decisions about recalculation of raw data values to unipolar scale.
This article focuses on a selection of the data measured in the Active Plus intervention and, as already mentioned, analyses are performed at concept-level. The selection consists of data about the main determinants of physical activity behaviour, including some social-related determinants, the main outcome measure from the SQUASH questionnaire and some variables indicating the intervention content the participant receives. As described, the intervention content that an individual participant has received is personalised and proxied in the analyses. The proxy of the intervention content is represented in the data by intervention condition variables, which thus play a central role in analyses. Table 1 gives an overview of these and all other concepts included in this article’s analyses, indicating the number of item-level variables the concept variable aggregates and at which moments in time the concept is measured. Note that the number of items for the SQUASH outcome measure is not indicated since it is calculated by standard rules.
3.3 Missing Data Analysis
A significant part of this case study consists of the evaluation of several ways to handle missing data values. This subsection illustrates the magnitude of the missing data problem in the case study and determines which methods are appropriate to be evaluated.
A total of 39 variables being concepts at certain moments in time are selected as subset for analyses. Table 2 demonstrates the number of missing values out of 1976 observations for each of the included concept-level variable. Since the time dimension is crucial to analyse intervention effects and, as can be seen in Table 2, more than a fourth of the values are missing for measurements after the baseline, applying pairwise deletion would result in an immense loss of information. Furthermore, the number of complete observations is for the selection of concepts 360 out of 1976 in total, meaning that applying list-wise deletion would neglect a large part of the dataset. Since deletion methods are not appropriate to deal with the missing data in this case study, we resort to the remaining methods for dealing with missing data, i.e., mean imputation, multiple imputation and the SEM algorithm described in Sect. 2.3, are applied and results are compared.
3.4 Approach
This subsection discusses how a suitable method for handling missing data is determined in order to model the intervention data. To perform experiments, the bnlearn package in R is used for Bayesian network learning [10]. Source code has been made publicly availableFootnote 1.
In the comparison of the methods to handle missing data values evaluated in this article, we apply discrete dynamic Bayesian networks for preprocessed data that is discretised by manually creating intervals meaningful in the health psychology field. The models are learnt by the tabu search algorithm optimising the BIC score (see Sect. 2.2). Model parameters are learnt by the Bayesian method, where the imaginary sample size setting prevents zero probabilities in the conditional probability tables while keeping parameter estimates close to maximum likelihood estimates. In the intervention study at hand only system missing values occur, for example, in case a participant has not answered a specific question in the questionnaire or if the maximum amount of items allowed to be missing is exceeded. The methods evaluated apply imputation where missing values are substituted by (maximum likelihood) estimators during the structure learning phase, namely mean imputation, multiple imputation and the structural EM algorithm, introduced in Sect. 3.3. Different variants of multiple imputation are evaluated, imputing \(m = 3\), \(m = 10\) or \(m = 20\) datasets, where each variable in each dataset is imputed using a classification tree. The parameter m has been chosen to evaluate its effect on the performance of multiple imputation within acceptable bounds for evaluation running time. The m datasets are analysed by creating a fully directed averaged Bayesian network model. These methods are compared by means of comparing the mean test-set log-likelihood using k-fold cross-validation (with \(k=10\)).
Finally, a linear Gaussian temporal Bayesian network model for the Active Plus intervention data is constructed from the preprocessed selection of data by learning the network structure using SEM. The model is learnt by the tabu search algorithm, optimising the BIC score, and maximum likelihood parameters are learnt in the continuous case. It was chosen to learn a continuous network rather than a discrete one to prevent possible loss of information from the discretisation process. In order to evaluate significance of edges, a bootstrap analysis is applied. Edges that are identified in most bootstrap samples and in the original network are considered stable findings in the following.
4 Results
This section describes the performance comparison of the methods applied to handle missing values. Furthermore, the learnt Bayesian network to model the Active Plus data is presented and results are compared to previous analyses of relations between determinants in the study by Van Stralen et al. [13].
4.1 Comparison Bayesian Network Missing Data Strategy
Table 3 demonstrates the mean log-likelihood over the folds resulting from applying the implemented cross-validation algorithm to the selected methods for handling missing data.
The cross-validation analysis shows that the structural EM algorithm significantly outperforms mean imputation and multiple imputation to handle missing data, because of significant difference of the mean log-likelihoods over the folds at 5% confidence level. Note that increasing the number of imputed datasets in multiple imputation significantly improves the performance of this method, which significantly outperforms mean imputation, though only minor improvements are observed between \(m=10\) and \(m=20\), which suggests that it is close to convergence.
Based on these performance results of the evaluated methods, the structural EM algorithm is chosen to be applied in learning the Bayesian network model in this case study. In the next subsection, the learnt model is presented and results are compared to those from previous analyses.
4.2 Comparison of Bayesian Network Model to Previous Analyses
Figure 2 shows the union of the temporal Bayesian network model learnt by the tabu search algorithm and the result of bootstrapping (which we call averaged model). Note that the edges represent probabilistic dependencies that not necessarily imply causal relationships. Table 4 gives the summary statistics of the temporal Bayesian network model learnt and its averaged counterpart, indicating that model complexity is decreased in the averaged model. A comparison of these models shows that in the original temporal Bayesian network model learnt, 20.74% of the edges that occur are not present in the averaged model, represented by blue edges in Fig. 2. Besides, 12.35% of the edges appearing in the averaged model have not been selected in the original model learnt, which are represented by red edges in Fig. 2. These unstable edges should be analysed further in the future.
Compared to previous analyses, the Bayesian network model provides a more complete insight in the complexity of mechanisms influencing physical activity behaviour. Previously, mediation analyses by Van Stralen et al. [13] have shown that factors such as social modelling, self-efficacy and intention are significant mediators of the intervention influencing physical activity behaviour. In Fig. 3, a fragment of the stable part of the averaged model (Fig. 2) is shown that includes these previously proven significant determinants, intervention effects, and effects on physical activity. It also includes coefficients, which represent the maximum likelihood estimators of parameters of the Gaussian conditional density distribution of variables given their parents. This part of the network suggests that the intervention effect on physical activity levels is mainly mediated by influencing habit and intention. Since the concept of habit has not been included in analyses by Van Stralen et al. [13], no comparison can be made with respect to results about this concept. Note that results from the Bayesian network model in general would probably have been more comparable to those from previous analyses if this concept would have been included in both previous and this papers researches. Previous results with respect to the intention concept are confirmed. However, although the network confirms significant effects of social modelling and self-efficacy on physical activity, this model does not find a direct effect of the intervention on social modelling nor on self-efficacy. The direct influence of the intervention on these concepts previously found is thus not confirmed by the network model. This difference can be explained by looking at the whole Bayesian network model, where longer paths can be found from the intervention variable to these concepts via several other determinants. For example, the intervention influences intention, which is correlated with action planning that is again correlated with social modelling. The network thus indicates that important mediators of physical activity are rather indirectly influenced by the intervention.
The submodel in Fig. 3 further shows that the extension in which environmental components are added to the intervention does not significantly influence physical activity nor its determinants. This differs from results from previous analyses by Van Stralen et al. [13], where differences have been found between effects in groups of participants having received environmental content and those who did not receive this extension. The previously found significant influence of the environmental extension on physical activity and determinants is thus not confirmed by the Bayesian network, in case of focusing on stable edges. Taking into account unstable edges some correlations are found between the environmental extension variable and for example the commitment concept. The difference in results from the network model compared to previous findings with respect to the influence of environmental components might be explained in future analyses by exploring these unstable findings.
Furthermore, the submodel shows that a clear distinction can be made between determinants of physical activity in the short (T1) and in the long (T2 and T3) run. In the short run, effects on physical activity are mainly determined by self-efficacy and intrinsic motivation, which mediates effects of habit and self-efficacy measured at T0. In the long-run, social modelling, intention and habit are important, where habit at T2 has the strongest correlation with physical activity levels at T2. Recall that findings with respect to the habit concept cannot be compared to previous findings by Van Stralen et al. [13]. Previous analyses on mediator effects on physical activity, do have shown the significance of the other determinants of physical activity levels indicated by the network model, except for the intrinsic motivation concept that appears in the Bayesian network to significantly directly influence physical activity. The network model shows that the effect of self-efficacy on physical activity at T1 is both direct and mediated by intrinsic motivation, since self-efficacy at T0 influences intrinsic motivation at T0, which subsequently influences physical activity levels at T1 via intrinsic motivation levels at T1. In this way, the network model explores the mechanism in which self-efficacy influences physical activity. It shows, that intrinsic motivation emerges as mediator, since intrinsic motivation mediates effects of other determinants on physical activity, in this case self-efficacy. This result of intrinsic motivation being a significant determinant of physical activity in the short run is thus new, compared to results from previously performed classic mediator analyses. This new short run determinant is found by the network by revealing the structure in which other determinants, that have already been found in previous analyses, effect physical activity. This structure reveals concepts that mediate effects of previously found determinants.
5 Discussion and Conclusions
In this article, the Bayesian network modelling technique has been applied to an e-health intervention case study to achieve better understanding of relations between determinants of physical activity. The reason for that was that this technique has not been applied often in this field and traditional analyses are not sufficient to reveal the dependence structure between determinants.
One particular challenge has been the magnitude of the missing data problem, which is examined for this case study and shown to be of such size that conventional methods to handle it cannot be used. The performance of several suitable methods, i.e., mean imputation, multiple imputation applying classification trees and the structural EM algorithm, has therefor been evaluated. Results show that applying the structural EM algorithm leads to the best results in terms of goodness of fit when learning a Bayesian network model for intervention data. In particular, by evaluating different numbers of imputed datasets as parameter setting in the multiple imputation method, the results suggest that the multiple imputation method does not outperform the structural EM algorithm even if the number of imputed datasets is increased to a large number. Note that we have compared these methods using a single cross validation run. Repeated cross validation is often suggested to increase confidence in the estimates, though it has been shown [15] that applying repeated cross-validation does not necessarily give more precise estimates of model accuracy. Furthermore, since the differences are quite large, further investigation does not appear to be necessary.
Since the modelling technique of Bayesian networks has not yet often been applied in this research field, its added value compared to more classic analyses in health psychology is evaluated. The model learnt for the case study data, applying the, in cross-validation evaluated, best performing algorithm to handle missing values, shows that the intervention does influence physical activity behaviour. The model also confirms previously-found important mediators of these intervention effects. However, there is some room for improvement to increase confidence in relations in the model, due to some unstable edges found. These should be explored in future analyses and would hopefully clarify the added value of the environmental extension on intervention effects. In a submodel, including significant edges only, some differences with respect to intervention effects on determinants and mediation effects on physical activity have been found compared to previous analyses. The network model shows that some determinants are rather indirectly influenced by the intervention and reveals a new significant mediator of intervention effects on physical activity, as it mediates effects of another determinant.
By the design of the study, the extent to which the intervention aims to influence different determinants in content received at different moments in time, varies across the participants. Each participant receives a unique focus in content fitted to their personal characteristics. Since the (personalised) content of the intervention is not included in the dataset, i.e., it is a latent variable, some care should be taken in the interpretation of the results. In particular, when a certain concept appears in the Bayesian network to be a significant determinant of physical activity, then this suggests that it is important in determining this behaviour. However, we cannot exclude the possibility that the effect of this determinant on physical activity is only mediated by the (personalised) contents of the intervention. Therefore, for designing new interventions, care should be taken in interpreting the present results.
In conclusion, the network has provided a more in-depth view in the dependencies and the complex structure in which determinants and physical activity are influenced by the intervention. In this way, the added value of applying the Bayesian network model compared to traditional analyses has been shown as the model provides new information relevant to understand the working mechanisms of the intervention, though care should be taken in the interpretation of the results. Nonetheless, the results show that Bayesian networks provide a useful technique to better understand dependence mechanisms of determinants of behaviour change.
6 Future Work
In future work, analyses in this article could be extended for example by evaluating other imputation methods to be implemented in the structural EM algorithm, such as a distribution over values instead of imputing the value with highest probability (soft EM). Also, although imputation using classification trees in multiple imputation has been an informed choice, evaluating alternative methods would be interesting. From a technical perspective, we will also consider exploring constraint-based structure learning algorithms, other score-based algorithms, alternative parameter learning algorithms or alternative model selection criteria. From the application perspective, future research could further elaborate on the structure, in which determinants are related to each other and physical activity, and on the differences found in the Bayesian network model compared to previous (regression) analyses.
A combined model could in future be designed for an integrated dataset including measurements from several different e-health intervention studies. Data on different sub-populations could be combined in order to examine if a general model yields different or additional results compared to the submodels for a smaller amount of data from single studies. It would also be interesting to perform analyses on an integrated dataset in more detail by using item variables in order to clarify correlations between concepts in a network model learnt for concept variables.
Even with data from a single study, this paper already shows that the Bayesian network model provides a more complete and in-depth insight in dependency structures, by exploring differences between its results and those from previous analyses. More specifically, the network reveals relations between variables where a variable influences another via a third one. In previous analyses, only some of the hypothetical mediator effects are explored by regression analyses. Hence, our results provide new opportunities to analyse and confirm our findings using traditional statistical methods.
Change history
16 October 2021
“Gaining Insight into Determinants of Physical Activity Using Bayesian Network Learning” was previously published non-open access. It has now been changed to open access under a CC BY 4.0 license and the copyright holder updated to ‘The Author(s)’. The book has also been updated with this change.
References
Brug, J., van Assema, P., Lechner, L.: Gezondheidsvoorlichting en gedragsverandering, 9th edn. Koninklijke Van Gorcum, Assen (2017)
Dempster, A.P., Laird, N.M., Rubin, D.B.: Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. B (Method.) 39(1), 1–22 (1977)
Friedman, N.: The Bayesian structural EM algorithm. In: Proceedings of the Fourteenth Conference on Uncertainty in Artificial Intelligence, pp. 129–138 (1998)
Ji, Z., Xia, Q., Meng, G.: A review of parameter learning methods in Bayesian network. In: Huang, D.-S., Han, K. (eds.) ICIC 2015. LNCS (LNAI), vol. 9227, pp. 3–12. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-22053-6_1
Koller, D., Friedman, N.: Probabilistic Graphical Models: Principles and Techniques. MIT press, Cambridge (2009)
Murphy, K.: Dynamic Bayesian networks: representation, inference and learning. Ph.D. thesis, UC Berkeley (2002)
Nakai, M., Ke, W.: Review of the methods for handling missing data in longitudinal data analysis. Int. J. Math. Anal. 5(1), 1–13 (2011)
Pearl, J.: Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. Morgan Kaufmann, Burlington (1988)
Schwarz, G.: Estimating the dimension of a model. Ann. Stat. 6(2), 461–464 (1978)
Scutari, M.: Package ‘bnlearn’. Bayesian network structure learning, parameter learning and inference, R package version 4.4 1 (2019)
Scutari, M., Graafland, C.E., Gutiérrez, J.M.: Who learns better Bayesian network structures: accuracy and speed of structure learning algorithms. Int. J. Approximate Reasoning 115, 235–253 (2019)
van Stralen, M.M., Kok, G., de Vries, H., Mudde, A.N., Bolman, C., Lechner, L.: The active plus protocol: systematic development of two tailored physical activity interventions for older adults. BMC Public Health 8, 399 (2008)
van Stralen, M.M., de Vries, H., Bolman, C., Mudde, A.N., Lechner, L.: Exploring the efficacy and moderators of two computer-tailored physical activity interventions for older adults: a randomized controlled trial. Ann. Behav. Med. 39(2), 139–150 (2010)
van Stralen, M.M., de Vries, H., Mudde, A.N., Bolman, C., Lechner, L.: Determinants of initiation and maintenance of physical activity among older adults: a literature review. Health Psychol. Rev. 3, 147–207 (2009)
Vanwinckelen, G., Blockeel, H.: On estimating model accuracy with repeated cross-validation. In: Proceedings of the 21st Belgian-Dutch Conference on Machine Learning, pp. 39–44 (2012)
Wendel-Vos, G.C., Schuit, A.J., Saris, W.H., Kromhout, D.: Reproducibility and relative validity of the short questionnaire to assess health-enhancing physical activity. J. Clin. Epidemiol. 56, 1163–1169 (2003)
Acknowledgements
This work is part of the research programme Active4Life with project number 546003005, which is financed by ZonMw.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this paper
Cite this paper
Tummers, S.C.M.W., Hommersom, A., Lechner, L., Bolman, C., Bemelmans, R. (2021). Gaining Insight into Determinants of Physical Activity Using Bayesian Network Learning. In: Baratchi, M., Cao, L., Kosters, W.A., Lijffijt, J., van Rijn, J.N., Takes, F.W. (eds) Artificial Intelligence and Machine Learning. BNAIC/Benelearn 2020. Communications in Computer and Information Science, vol 1398. Springer, Cham. https://doi.org/10.1007/978-3-030-76640-5_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-76640-5_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-76639-9
Online ISBN: 978-3-030-76640-5
eBook Packages: Computer ScienceComputer Science (R0)