Abstract
Subaerial biofilm (SAB) formation on cultural heritage objects is often considered an undesirable process in which microorganisms and their by-products, e.g., enzymes and pigments, cause damage or alteration to a surface. Since biofilms are widespread phenomena, there has been a high demand for preventive and control strategies that resist their formation or reduce their negative effects once formed. Up to date, the main strategy to control biofilms has been the use of biocides. Because of their intrinsic properties, biocidal products can pose risks to humans, animals, and the environment. In this chapter, the authors call “green” only those alternative strategies to biocides able to prevent/control biofilms but that do not kill microorganisms, i.e., irrespective of the use of natural compounds. Here, we describe some of the methods that are most commonly used to test the effectiveness of antibiofilm compounds with multiple-species biofilm model systems. A unified terminology and well described protocols and guidelines are still required to compare and test the effectiveness of traditional or novel compounds against biofilms retrieved on heritage surfaces.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Biocides
Biofilms are highly structured communities of microbial cells that adhere to a surface and are embedded in extracellular polymeric substances (EPS) consisting of polysaccharides, nucleic acids, proteins, and lipids. Biofilm is a natural and ubiquitous form of bacterial and fungal growth. The fact that EPS are commonly observed close to their cells also suggests that lichens are biofilms (Banfield et al. 1999; De Los Rios et al. 2005; Grube and Berg 2009; Casano et al. 2015). The concept of the biofilm implies a need to take into consideration the interactions established among microorganisms, and of the microorganisms with the heritage substratum and the immediate environment, a view that overcomes the value-laden identification of microorganisms and their activities as single taxon.
In the scientific literature, biofilms have been mainly investigated when growing on a solid surface and exposed to a liquid phase. Indeed, most studies have focused on human infecting biofilms, typically at the solid–liquid interface of the inner body and in marine biofouling environments. Medical literature addressing the solid–air interface is considerably less substantial and concerns (partially) only skin wound biofilms. Despite many similarities between biofilms grown at a liquid–solid interface and at an air–solid interface, there are marked differences that have been noted only relatively recently. In contrast to the solid–air interface, Bacillus subtilis forms sessile pellicles at liquid–air and solid–liquid interfaces (Vlamakis et al. 2013). In this scenario, the work of cultural heritage microbiologists that mainly addresses sessile life at solid–air interfaces is pivotal to study a biofilm aspect that is less investigated.
Subaerial biofilm (SAB) formation on cultural heritage objects is often considered an undesirable process in which microorganisms and their by-products, e.g., enzymes and pigments, cause damage or alteration to a surface. Since biofilms are widespread phenomena, there has been a high demand for preventive and control strategies that resist their formation or reduce their negative effects once formed. Up to date, the main strategy to control biofilms has been the use of biocides. According to the European Regulation 528/2012, “biocidal product” means “any substance or mixture, in the form in which it is supplied to the user, consisting of, containing or generating one or more active substances, with the intention of destroying, deterring, rendering harmless, preventing the action of, or otherwise exerting a controlling effect on, any harmful organism by any means other than mere physical or mechanical action” (Regulation (EU) No 528/2012 of the European Parliament and of the Council of 22 May 2012). In the past, biocides have been freely used to control biofilms on cultural heritage surfaces. However, the use of biocides has been proven to have major drawbacks: biocides harm non-target populations of the surrounding environment and they carry a risk of the development of resistance to themselves as well as cross-resistance to antibiotics (Villa et al. 2020). Thus, because of their intrinsic properties, biocidal products can pose risks to humans, animals, and the environment. As a result, the EU has set up strict rules and procedures to minimize these risks and all biocidal products have to be authorized by a competent authority before they are placed on the market (https://ec.europa.eu/health/biocides/overview_en).
Moreover, specifically in relation to conservation, before deciding whether removal of microbial colonization with biocides is actually beneficial, we should consider the effects of this so-called biodeterioration in comparison with the physico-chemical deterioration induced by the use of the biocidal agent. In outdoor conditions, once structures and objects have been cleaned, recolonization is inevitable and may begin shortly after treatment. In many cases no well-defined differences in the microbial community between the untreated and the treated surfaces are seen as only a biofilm component is affected (Urzì et al. 2016). In other cases, subsequent colonization might be more aggressive or disfiguring than the one eradicated (Martin-Sanchez et al. 2012).
In conservation as well as in all other fields, the new approach is to find more sustainable ways to use biocides for the removal of deleterious biofilms from relevant abiotic surfaces and, at the same time, to search for feasible sound alternatives.
2 What Do We Mean by Green Alternatives?
In this chapter we define green alternatives those able to prevent/control biofilms without affecting cell growth. One of the many advantages is that, if the antibiofilm effect is not due to a biocidal effect, it is possible to prevent the generation of microbial drug-resistance.
At first glance, physical techniques seem a good green alternative for the removal of biodeteriogens. These methods dislodge or cause the lysis of some microorganisms with the help of tools, such as brushes (Caneva et al. 1991; Borderie et al. 2015). In contrast to biocides, the killing does not lead to resistance but the material can be discolored or damaged (López et al. 2010; Vujcic et al. 2019). Additionally, the effect is mainly on the surface, e.g., endolithic growth is generally not affected. Therefore, physical cleaning is not the focus of this chapter even if its use in combination with other methods discussed here can be considered.
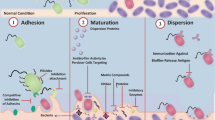
Antibiofilm agents target various stages of biofilm growth, namely adhesion, maturation, and dispersion. Any action that inhibits adhesion is considered a preventive approach, whereas any action that interferes with maturation and dispersion is included in a control strategy. Prevention can be operated with two strategies: either through the employment of specific substances in the repository environment or by modifying the surface of the heritage object. For instance, essential oils in the vapor phase or blended with some solvent have the potential to be used as antibiofilm substances and can be considered an interesting alternative (Borrego et al. 2012).
From an ecological point of view, the selection of plants as source materials of antimicrobial compounds is a good approach, since plants produce a wide range of secondary metabolites that naturally defend them against microorganisms. Silva et al. (2016) reported a comprehensive list of plant metabolites that have been proven effective against microorganisms and their products, including pigments. We can argue about whether this is the green alternative we need. “Natural,” including plant-derived compounds, is not a synonym of non-toxic or non-biocidal. In the past, thymol vapor was used extensively by a number of conservators in “thymol cabinets.” However, in respect to heritage materials thymol softens varnishes and resins, renders parchment brittle, degrades some paper supports, watercolor binders, and iron gall ink, and, with respect to human health, librarians and archivists suspected it to be a carcinogenic substance (Holben Ellis 1995; Isbell 1997). Indeed, the conclusion of the peer review of the pesticide risk assessment of the active substance thymol by the European Food Safety Authority, reached on the basis of the evaluation of the representative uses of thymol as a fungicide on table and wine grapes, reported that “a high risk was identified for aquatic organisms, leading to a critical area of concern” and “the formulated product contains the impurity methyleugenol which is a genotoxic carcinogen” (EFSA 2012).
The use of alternative substances can be considered after making all the preliminary tests to prove the worth of them. The first step is to evaluate the threshold above which the compounds inhibit microbial growth. The sixteenth century scientist Paracelsus was the first to state that all things are poisons, and that the degree of toxicity is only caused by the dose. In microbiology, the minimum inhibitory concentration (MIC) identifies the minimum amount of the compound that is required to inhibit microbial growth, under defined laboratory conditions. MIC has been used for decades and measures the concentration required to inhibit growth or to kill planktonic microorganisms. Importantly, it is now known that for some substances, the resistance of biofilm bacteria may be a thousand times greater than that of planktonic bacteria of the same strain (Olson et al. 2002). Studying the photocatalytic titanium dioxide (TiO2) nanopowder and TiO2 thin film, Polo et al. (2011) showed one order of magnitude reduction of Pseudomonas aeruginosa planktonic cells in 2 h and an almost complete eradication of P. aeruginosa planktonic cells, respectively. In contrast, neither the photocatalytic treatment with TiO2 film nor that with TiO2 nanopowder had any effect on P. aeruginosa biofilms. Nevertheless, when studying non-biocidal antibiofilm substances it is possible to use MIC. In fact, if the selected compound does not kill planktonic cells at the concentration adopted, it is even less likely that it will kill sessile cells.
Before use, the environmentally friendly organic compounds have to be proven not to act as carbon and energy sources for the target microorganisms. The rationale of the experiment is that, if a substance does not kill a microorganism, it can function as a nutrient. To investigate this matter the substance can be supplied as the only carbon and energy source for the target microorganisms in various amounts.
Another concern of using phytochemicals, such as oils and plant extracts, is that their composition varies over time. For instance, Nezhadali et al. (2014) claimed that the composition and quantity of essential oil from a specific thyme species can be considerably influenced by harvesting season, geographical location, and other agronomic factors. Additionally, based upon soil type variations, distinct differences among chemotypes can be found over a few meters. Moreover, extrinsic factors related to the extraction method affect their chemical composition (Dhifi et al. 2016). The apparently different results of the compounds once extracted from the same plant sources often found in literature are easily explained by the fact that the tested materials have a different composition. Consequently, all these plant-derived mixtures must be chemically characterized or otherwise other researchers cannot repeat the proposed experiments. Otherwise chemically synthetized compounds can be purchased from the market. In this respect, it is also worth noting that some phytochemicals, such as essential oils, are very expensive. Therefore, a valuable alternative to essential oils is to use the main pure synthetic counterpart that is also generally more stable (Rakotonirainy and Lavédrine 2005). Indeed the use of essential oils has been proposed in conservation literature as a “green alternative” (Macro et al. 2018). However, in this chapter, although they can be effective, we do not define as a green approach any alternative to common biocides that uses products of natural origin but we define green a strategy that does not kill microorganisms.
Disrupting the biofilm not killing the cells is not yet a reality in conservation. In this line of thought, no data are currently provided on the effectiveness of non-biocidal strategies as alternatives to control biofilms. However, at present, it is possible to suggest multiple approaches in addition to biocides in order to reach an effective clearance of biofilms without a massive use of toxic substances. This has been proven with nitric oxide (NO). NO is a signaling molecule involved in the modulation of quorum sensing (QS), a method of cell-to-cell communication, able to elicit bacterial dispersal (Kyi et al. 2014). Microorganisms isolated from the biodeteriorated wooden sculpture So It’s Come To This (1986) by Bruce Armstrong, at the University of Melbourne headquarters, have been treated with a nitroxide, a compound of which the antibiofilm mechanism is similar to that of NO, while it is less expensive and with a longer life than NO-donors (Alexander et al. 2015; Alexander and Schiesser 2017). A 24 h treatment with 50 μM nitroxide followed by 2 h treatment with 0.001% w/v benzalkonium chloride effectively eradicated biofilms. Importantly, in this study, the biocide was used at a concentration much lower than those usually employed (2% w/v).
In regard to microbiological issues, enzymes have been employed in conservation for removing microbial staining (Konkol et al. 2009), for monitoring biodeterioration (Rosado et al. 2013), and for killing microbial cells (Valentini et al. 2010). In the Chinese literature (Wu and Lou 2016), chitinases were also used to inhibit the development of filamentous fungi growing on word walls and canoes at the Cross Lake Bridge ruins in Xiaoshan. However, the authors of this chapter could not understand in depth this research (in particular, whether toxicity was evaluated and fungal biofilm was formed) as only the abstract of the Chinese manuscript has been translated into English. Another application with the potential for green antibiofilm technology is the use of enzymes to degrade the extracellular matrix for biofilm dispersal, including glycosidases, proteases, and deoxyribonucleases (Kaplan 2010; Shadia and Aeron 2014), also immobilized to a surface (Spadoni Andreani et al. 2017). To the best of our knowledge, matrix-degrading enzymes have never been tested to disperse biofilms in the conservation field but EPS inhibitors, such as a mixture of bismuth nitrate and dimercaprol, have been successfully used during the EU project BIODAM (Robertson et al. 2004).
Metal cations, such as calcium, magnesium, and iron have been implicated in maintaining matrix integrity. Antibiofilm formulations incorporating ethylenediaminetetraacetic acid (EDTA) and other permeabilizers have shown efficacy on in vitro biofilms in synergism with antimicrobial agents (Robertson et al. 2004). Nuclear fast red and methylene blue, two photodynamic agents investigated in combination with hydrogen peroxide in the EU project BIODAM (Young et al. 2008), showed the potential to destroy cyanobacteria on stone samples and, since photodynamic agents are themselves subsequently degraded by visible light, the substratum is not discolored. Neither light alone nor the presence of H2O2 led any change in the fluorescence of Synechococcus leopoliensis. Combining methylene blue with hydrogen peroxide resulted in a decrease of the fluorescence of 40%.
Interestingly to mention in this chapter, even if some natural products were selected with the aim to kill the target microorganisms they showed much lower toxicity in respect to non-target microorganisms in comparison with traditional biocides. This is the case of metabolites produced by Bacillus spp. that showed no lethality against brine shrimp and Swiss mice through administration of 5000 mg/kg acute dose (Silva et al. 2016b). In contrast, Preventol® caused acute toxicity with 10 times minor concentration dose administrated in the same conditions.
Unfortunately, at present, only few molecular mechanisms of action related to some antibiofilm agents are known. Proteomic analysis of biofilm exposed to the antibiofilm zosteric acid sodium salt and salicylic acid revealed that a number of proteins were up- and downregulated, and these proteins were associated with stress, motility, cell-to-cell communication, reactive oxygen species accumulation and metabolism (Villa et al. 2012; Cattò et al. 2017). Recently, the zosteric acid sodium salt and the usnic acid have been encapsulated in silica nanosystems commonly used to protect stone surfaces (Ruggiero et al. 2020). The antifouling activity was successfully assessed against planktonic microorganisms from biopatinas colonizing the Aurelian Walls in Rome. Hopefully, in the near future, the above coatings will be tested in situ and their antibiofilm properties will be investigated.
3 Lab Biofilm Systems to Test the Efficacy of an Antibiofilm Compound/Mixture
On the one hand, the use of typical laboratory planktonic microorganisms for selection of biocides is inappropriate. On the other hand, biofilms on cultural heritage surfaces are inherently heterogeneous. Consequently, researchers have made use of laboratory biofilm models that capture the salient features of target sessile cells while providing numerous and consistent biofilm samples. Therefore, first of all, a biofilm at the solid–air interface (SAB) has to be formed.
In fact, while field experiments are clearly instrumental to evaluate the efficacy of an antibiofilm compound, their design and execution face several challenges: small sample size, unrepeatable samples, and complex sample structure. Furthermore, many field-based studies are time-limited as many biofilm processes relevant to the performance evaluation of antibiofilm agents occur over a very long time, for example, processes such as biofilm regrowth and succession as well as response to various environmental conditions.
The model systems are in contrast with field systems in terms of simplicity and accessibility. The purpose of a laboratory model system is not to represent a mini version of field systems, but rather to simplify nature so its processes can be easily understood (Jessup et al. 2004).
In the biofilm community it has long been debated whether a bacterial colony on a solid growth medium is to be considered a biofilm. Definitely, few colony morphologies can effectively reflect some of the important attributes of biofilms (Haussler and Fuqua 2013). However, as this question is not convincingly answered yet and, in conservation, a solid surface is better for mimicking a cultural heritage substratum, other systems have been considered better performing to achieve this aim.
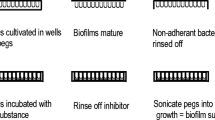
A colony biofilm is grown on a semipermeable membrane that sits on an agarized medium (Fig. 5.1). The membrane is a solid interface and it is semipermeable to allow nutrients to reach the cells. This method has been especially devised in the study of the antibiotic-resistant properties of cells (Merritt et al. 2005). Sessile cells can be given a new supply of nutrients by relocating the membrane with biofilm on a fresh agar plate. One can therefore easily assess the effect of the antibiofilm compound at the time of adhesion (inoculum) or when the biofilm is already mature. The membrane can be also modified for binding antibiofilm agents. The colony biofilms can grow quickly, are easy to handle, and require inexpensive laboratory materials. Recently, protocols for the growth of cyanobacterial and fungal colony biofilms have been set up by Sanmartín et al. (2015) and Gambino et al. (2017), respectively.
Biofilms that are composed of a mono-species are relatively rare in nature; rather, microorganisms are generally found in complex multispecies communities. In line with this observation, Miller et al. (2008, 2009) cultivated on lithic surfaces a natural green biofilm from an enriched microbial consortium residing on a limestone monument. However, the main question is how the original community adapts and evolves under the artificial conditions created in the laboratory systems. Recently, Vázquez-Nion et al. (2016) developed a multispecies liquid culture from natural SABs inhabiting granitic historic buildings, with the aim to use it as the inoculum for lab-scale biofilms. The researchers observed that after one year, the microbial community of the culture was stable and dominated by members of the Chlorophyta and Cyanobacteria, which were not part of the core microbiome of the original SABs.
Furthermore, natural biofilm communities are difficult to analyze at molecular level. This may limit the comprehension of the mechanism of antibiofilm compounds, since omics-based technologies ideally require microorganisms with available genetic and physiological information (Noack-Schönmann et al. 2014).
To overcome the limitations arising from the complex phototrophic SABs, elegant dual-species biofilm models based on phototroph–heterotroph associations were developed, starting from two species that do not originate from an environmental microbial consortium and have never been introduced to each other.
A model system comprising the cyanobacterium Nostoc punctiforme strain ATCC 29133 (PCC 73102) as phototroph, and the well-studied marble-derived microcolonial fungus A95 Knufia petricola (syn. Sarcinomyces petricola) as the heterotrophic component was developed by Gorbushina and Broughton (2009). This model allowed Seiffert et al. (2014, 2016) to successfully study the biological impact of the consortium on weathering granite and related minerals in a geomicrobiologically modified percolation column. This system is promising for testing antibiofilm agents, because the compound can percolate into the column and interact with the biofilm. At the end of the treatment, the biofilm biomass can be removed from the system for a further analysis.
Villa et al. (2015) used a laboratory model of SABs composed of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 and the chemoheterotroph Escherichia coli K12 (Fig. 5.2b). A modified drip flow reactor (DFR) was used to mimic a monument surface (Fig. 5.2a). SABs on outdoor stone monuments follow the water flow downward, experiencing low fluid velocity over the surface, while the biomass is continuously exposed to the air. Similarly, biofilms in the DFR are under low-shear/laminar flow as the medium drips on a 10-degree surface, and a high gas transfer environment as the biofilm is continuously exposed to the air in the head space. This setup was effective in reproducing SABs as it was able to capture typical features of biofilms on outdoor stone monuments such as: (i) microcolonies of aggregated bacteria; (ii) a network-like structure following surface topography; (iii) cooperation between phototrophs, heterotrophs, and cross-feeding processes; (iv) ability to change the chemical parameters that characterize the microhabitats, and (v) survival under desiccation stress. By using the SAB model system, Villa et al. (2015) compared the susceptibility of biofilm and planktonic cells to a disinfection treatment based on a quaternary ammonium salt. The results revealed that the biofilm retained more viable cells than the planktonic cells after 45 min of antimicrobial exposure. These results suggested that the dual-species SAB exhibits an antimicrobial tolerance, which is the hallmark of the biofilm mode of growth.
Panel (a) is the schematic representation of the modified drip flow reactor (DFR) used by Villa et al. (2015). Panel (b) displays a picture of DFRs with a glass lid (panel b). Panel (c) shows the 3D reconstruction of dual-species SABs grown in the DFR following the method reported by Villa et al. (2015). Color legend: E. coli cells are green (GFP); Synechocystis cells are red (autofluorescence); a reflection from inorganic materials is gray
While SAB models on stone are well developed, few lab-scale systems are available for other heritage substrata. Rakotonirainy and Lavédrine (2005) designed a system to reproduce fungal SABs on paper (Fig. 5.3). At the beginning a piece of paper is inoculated with a single strain or a mixture of many strains, letting the mycelium cover the entire surface and dry before inserting it in a book. Then the closed books are placed some centimeters high on a grid in a transparent test-chamber (Rakotonirainy and Lavédrine 2005). The tested antibiofilm agent impregnates a paper disc in an open plate on the bottom of the chamber. Humidity and temperature are set at the desired level. After exposure of the biofilms on paper to the vapor of the antibiofilm for a specific time cells can be investigated. Rakotonirainy and Lavédrine (2005) demonstrated that, at the concentrations tested, linalool, the main component in lavender oil, is fungistatic and could be employed in preventing fungal growth. Interestingly, this compound cannot be used as a fungicide as higher concentrations lead to a decrease in paper pH.
Schematic representation of the system designed by Rakotonirainy and Lavédrine (2005) to reproduce biofilms on paper
4 Methods for Testing the Effectiveness of Antibiofilm Substances
Despite the vast use of traditional and novel antibiofilm agents in conservation of cultural heritage, these practices are being increasingly questioned because of the lack of correlation between results obtained with conventional susceptibility tests at laboratory-scale and those obtained from in-field applications. The reason for this discrepancy is based on the use of planktonic cells for testing antibiofilm at laboratory-scale rather than sessile cells. As previously mentioned, it is now well known that biofilms are ∼1000-fold more resistant than their planktonic counterparts (Hall and Mah 2017). Furthermore, the lack of simple reliable and standard assays has remained a major limitation in selecting adequate antibiofilm treatments.
In view of the previous considerations, biofilm model systems are instrumental for testing the effectiveness of antibiofilm substances before an application in-field. Once the biofilm is grown, it can be exposed to different treatments and environmental conditions. Furthermore, biofilm model systems can be used to study biofilm regrowth when the treatment is concluded.
The antibiofilm agents can affect biofilm biomass, impair the biofilm matrix, or affect biofilm thickness and morphology (e.g., inability of fungi to form filamentous structures), while cells remain metabolically active. Thus, several methods can be applied to evaluate the impacts of a compound against the biofilm. Here, we briefly describe some of the methods that are most commonly used to test the effectiveness of antibiofilm compounds.
4.1 Plate Count Assay
To be able to assess the effectiveness of antibiofilm compounds, it is essential to determine the amount of viable cells in the biofilm, or the relative reduction in viable cells after the treatment. The most widely used technique to determining bacterial viability is counting the number of colony forming units (CFU) after plating cultures on agar media. This method to assess biofilm viability requires the dislodgment of the biomass from the surface and the disaggregation of cell clusters due to the presence of the biofilm matrix. Procedures such as vortexing, sonication, or matrix-dissolving enzymes can be applied to separate bacterial cells from the matrix. However, these procedures may affect bacterial viability, leading to significant errors in the quantification of the biomass (Azeredo et al. 2017). Furthermore, many strains cannot be cultured, as well as metabolically inactive cells (such as dormant cells and spores) commonly present within the biofilm (Sebestyen et al. 2016). Finally, while CFU counting can be a valuable tool for quantifying a bacterial biofilm, it is not suitable for fungal biofilms that develop filamentous structures and spores.
4.2 XTT Cell Viability Assay
The tetrazolium salt assay (XTT assay) is another useful technique for assessing the viability of biofilm cells in vitro, as well as their susceptibility to antimicrobials. The number of viable biofilm cells can be deduced by measuring the absorbance of the supernatant after the metabolic reduction of XTT into the water-soluble formazan (Pantanella et al. 2013). Since the formazan product is water soluble and easily measured in cellular supernatants, the XTT assay allows the investigation of biofilm agent susceptibility without disruption of the biofilm structure (Kuhn et al. 2003). The main limitation of this method is related to the biofilm heterogeneity with gradients of substrata and products, which lead to cells with different metabolic states. Furthermore, the biofilm matrix might slow down the reduction of XTT or partially retain and release the formazan.
4.3 ATP-Bioluminescence Assay
The bioluminescence assay, based on the use of the North American firefly luciferase, sees the conversion of intracellular ATP, the main energy carrier in cells of all living organisms, into light. The light generated is recorded by a luminometer and quantified in relative light units (Nante et al. 2017). Intracellular ATP content is a good indicator of cell viability. In fact, upon cell death, ATP synthesis is immediately arrested, while its hydrolysis can continue for some time. The main drawback of this method is related to the variation in ATP production among diverse microbial taxa (Ivanova et al. 2006).
4.4 Spectrofluorometric Assay
Fluorescein diacetate (3′,6′-diacetyl-fluorescein, FDA) is a pre-fluorophore that can be hydrolyzed by a wide spectrum of nonspecific extracellular and membrane-bound enzymes. Once hydrolyzed, FDA is converted to fluorescein that shows a yellow-green color absorbing at 490 nm. The method generally assumes the determination of FDA-hydrolyzing enzyme activity of cells that are detached from the carrier. Konkol et al. (2010) reported the use of beta-N-acetylhexosaminidase activity as a rapid and reliable means of fungal detection on a variety of cultural heritage materials. Adapted for use on cultural heritage materials, fluorogenic 4-methylumbelliferyl (MUF) labeled substrate N-acetyl-beta-d-glucosaminide (NAG) was used to detect beta-N-acetylhexosaminidase activity in fungi that were actively growing on library materials (Konkol et al. 2010). Being a metabolic assay, a significant limitation is the fact that the microorganisms in biofilms do not all display the same metabolic activity. Moreover, metabolic fluorimetric-based assays are often calibrated against planktonic cultures, introducing significant error as the metabolic rates differ greatly between the planktonic and the biofilm states (Welch et al. 2012).
4.5 Real-Time Reverse Transcription PCR (qRT-PCR)
Real-time quantitative-reverse transcription PCR (qRT-PCR) is one of the most powerful and sensitive gene analysis techniques available so far, in which the fluorescent signal is measured in real-time at each amplification cycle and is directly proportional to the number of amplicons generated. Moreover, since qRT-PCR detects mRNA, with its short half-life, it is a promising indicator of cell viability (Guilbaud et al. 2005; Xie et al. 2011). Thus, qRT-PCR can be applied not only to detect but also to quantify viable microorganisms in the biofilm (Ettenauer et al. 2015; Piñar et al. 2015). The main limitations are related to sample preparation, including the extraction of RNA.
4.6 Chlorophyll
Chlorophyll-a is a good estimator for quantifying microalgal and cyanobacterial biofilm biomass (Jesus et al. 2006; Sanmartín et al. 2010; Sendersky et al. 2017; Chamizo et al. 2018). The method requires the dislodgment of the biomass from the surface and the extraction of chlorophyll-a in DMSO followed by spectrophotometric measurements (Fernandez-Silva et al. 2011). Alternatively, biofilm biomass measurement can be achieved by measuring chlorophyll fluorescence. Recently, pulse amplitude modulated (PAM) fluorometry was used by Vázquez-Nion et al. (2018) to measure in vivo the fluorescence signal in a non-destructive way. The authors developed an indicator of fitness for photosynthetic organisms by quantifying the maximum photochemical efficiency of photosystem II. To this end, they measured the minimal fluorescence signal of dark-adapted cells and the maximal fluorescence signal after a saturating light pulse in dark-adapted cells and they developed a standard curve that allowed the correlation of the minimal fluorescence signal of dark-adapted cells with the amount of chl-a content as a biofilm biomass estimator.
4.7 Proteins Quantification
Protein cellular content can be used to estimate the total biofilm biomass. After cellular lysis, the released proteins can be quantified by colorimetric assays such as quantification, including the Bradford, Lowry, and bicinchoninic acid (BCA) methods. The most used method is the Bradford assay, which consists of adding an acidic Bradford reagent containing Coomassie Brilliant Blue G-250 dye to the lysed sample. Once the protein binds to the dye, changing the color from brown to blue occurs. The change in absorbance at 595 nm is recorded and converted to a total protein concentration through a standard curve (Bradford 1976).
4.8 Biofilm Staining and Microscopy
One of the most important advances in the study of biofilm has been the ability to visualize the effects of antibiofilm compounds on hydrated living biofilms in three dimensions, over time, using confocal laser scanning microscopy (CLSM).
The use of fluorescent probes allows imaging by multiple fluorochromes for evaluating the impacts of a compound on individual biofilm components simultaneously. Thus, specific fluorescent stains are also used in combination with CLSM for imaging biofilm cellular and extracellular matrix material. Furthermore, the intense development of specialized image processing software with user-friendly interfaces and the implementation of advanced CLSM techniques such as fluorescence lifetime imaging (FLIM), fluorescence correlation spectroscopy (FCS), and fluorescence recovery after photobleaching (FRAP) in commercially available confocal microscopes have improved the development of quantitative methods for the analyses of biofilm images.
The fluorescent stains are generally designed to bind a specific cellular component, such as DNA (e.g., propidium iodide, SYBR-green, and ToTo-1) or protein (e.g., Sypro-Ruby, 3-(4-carboxybenzoyl) quinoline-2-carboxaldehyde (CBQCA) or the NanoOrange) (Cattò and Cappitelli 2019). Furthermore, the intrinsic natural autofluorescence of some phototrophic microorganisms can be exploited for imaging differentiation. A comprehensive list of fluorescent stains commonly used in biofilm research has been provided by Neu and Lawrence (2014).
The LIVE/DEAD BacLight assay is a dual staining kit composed of two fluorophores, namely SYTO9 and propidium iodide (PI). The green-fluorescent nucleic acid stain SYTO9 enters live and dead bacterial cells and binds to DNA of both Gram-positive and Gram-negative bacteria. The red-fluorescent nucleic acid stain PI intercalates to DNA and it is commonly used for identifying dead cells in a population and as a counterstain in multicolor fluorescent techniques because it is supposed to penetrate only cells with disrupted membranes and it is generally excluded from viable cells. Thus, the SYTO9/PI combination allows one to detect bacterial viability based on the detection of membrane integrity. The main advantages of using the LIVE/DEAD BacLight assay are related to the rapid procedure, the possibility to perform quantitative analyses, as well as to measure the fluorescent signal using various instruments such as flow cytometers and microplate readers. The principal drawback of this method is the need to observe a statistically relevant portion of the sample to obtain representative information on the total population.
The coupling of CLSM with fluorescence in situ hybridization (FISH) probes leads to more information about the identification and localization of specific microbial taxa before and after a biocide treatment. In fact, FISH-CLSM allows microorganisms to be specifically labeled within the 3D extracellular matrix by using non-invasive imaging of fully hydrated biofilms. In short, biofilm cells are fixed, permeabilized to facilitate access of the probe to the target site and then hybridized with nucleic acid probes labeled with a fluorochrome. The classic FISH technique relied on ribosomal RNA as probe target, and thus it is traditionally applied for the phylogenetic identification of microorganisms in mixed assemblages without prior cultivation. Since the FISH signal is related to the cellular rRNA content, which reflects the cell activity, this technique can be used to evaluate the biocidal effect of a compound or a treatment towards target microorganisms.
Despite these promising features, the classic FISH protocol might generate weak fluorescent signals in metabolically active cells. This drawback can be attributed to the biofilm matrix that hinders the access of the probes into the cell, as well as to the low ribosome content in slowly growing or metabolically inactive biofilm cells. To overcome these limitations, different ways for increasing the FISH signal have been developed, including the use of multiple probes for one target microorganism, the use of peptide nucleic acid probes (PNA-FISH), and the use of catalyzed reporter deposition (CARD-FISH), just to name a few. Recently, a method based on combinatorial labeling and spectral imaging FISH (CLASI-FISH) has been developed to detect hundreds of different microbial taxa in single microscopy imaging (Valm et al. 2011; Behnam et al. 2012; Valm et al. 2012). Another disadvantage of FISH approaches is that the samples have to be processed with several treatments prior to the probe hybridization, which may disturb the structure of the biofilm and makes time-course studies difficult.
Most fluorescent dyes used in biofilm studies stain cellular components. Visualizing the extracellular matrix by CSLM has been more challenging. A few fluorescent stains are becoming available for CSLM studies of biofilm matrix components. In particular, fluorescent lectins bind to specific sugars and can be used to stain certain extracellular polysaccharides. Since lectins are large molecules, they do not penetrate biofilms well. Ideally, small molecule stains will become available for staining the polysaccharide component of biofilms, such as Calcofluor, which is used to stain biofilms of strains that produce cellulose (Spiers et al. 2003). Another component of the biofilm extracellular matrix material is extracellular DNA. Various stains are available that bind DNA, with the TOTO-1 iodide stain providing excellent contrast between the biofilm eDNA component and the biofilm cells (Gloag et al. 2013). By comparing the biovolumes of the matrix components before and after the treatments, it is possible to evaluate the effectiveness of an antibiofilm compound.
In addition to staining biofilm components, fluorescent stains are available for studying metabolic activities within biofilms. For example, tetrazolium salts precipitate when reduced by the biofilm cells, forming a zone of fluorescence around the active cells (Franklin et al. 2015). Calcein AM is a fluorogenic, cell-permeant fluorescent probe that indicates cellular viability in biofilms. The probe is nonfluorescent until acted upon by nonspecific esterases present in live cells. Thus, cleaving the AM ester allows the probe to emit a fluorescent signal that is proportional to cell vitality.
Calcein AM green stain has also been used to observe the permeabilization of biofilm cells by a biocide with time-lapse CLSM (Davison et al. 2010). The technique allowed for the simultaneous imaging of changes in biofilm structure and disruption of cellular membrane integrity through the loss of the intracellular green signal, generated by the cleavage of the AM ester prior to the antimicrobial exposure.
Daddi Oubekka et al. (2012) used a set of advanced fluorescence microscopic tools such as fluorescence recovery after photobleaching, fluorescence correlation spectroscopy, and fluorescence lifetime imaging, to characterize the dynamics of fluorescently labeled vancomycin in biofilms.
Thus, imaging techniques can be applied to elucidate the effectiveness of a biocide treatment by watching, through a microscope, the antimicrobial attack.
5 Conclusion
Biofilms are the dominant lifestyle of microorganisms in all environments, either natural or manmade, including heritage. The development of effective strategies to combat biofilms is a challenging task.
These emerging novel antibiofilm strategies are still in the nascent phase of development, and more research is urgently needed to validate these approaches, which may eventually lead to effective prevention and control of biofilms. Until now, the research and application of antibiofilm compounds have often been questioned owing to the diversity of the testing methods available and the variations of the results reported in the literature vs those obtained in-field. Thus, numerous innovative antibiofilm approaches have been published, but it is difficult to reliably compare all these strategies.
Some factors still hamper the testing and screening of antibiofilm compound such as, among others, the scarcity of homogenized testing protocols, the lack of normalized vocabulary, the difficulty of testing repeatability and reproducibly. Thus, a key aspect of future antibiofilm research is the need for standards: a unified terminology and well described protocols and guidelines are required to test the effectiveness of traditional or novel compounds against biofilms retrieved on heritage surfaces. These protocols and guidelines should be a preliminary step in the direction of a potential code of green practices.
References
Alexander S-A, Schiesser CH (2017) Heteroorganic molecules and bacterial biofilms: controlling biodeterioration of cultural heritage. Arkivoc part ii: 180–222
Alexander S-A, Rouse EM, White JM, Tse N, Kyi C, Schiesser CH (2015) Controlling biofilms on cultural materials: the role of 3-(dodecane-1-thiyl)-4-(hydroxymethyl)-2, 2, 5, 5-tetramethyl-1-pyrrolinoxyl. Chem Commun 51:3355–3358
Azeredo J, Azevedo NF, Briandet R, Cerca N, Coenye T, Costa AR, Desvaux M, Di Bonaventura G, Hébraud M, Jaglic Z, Kačániová M, Knøchel S, Lourenço A, Mergulhão F, Meyer RL, Nychas G, Simões M, Tresse O, Sternberg C (2017) Critical review on biofilm methods. Crit Rev Microbiol 43:313–351
Banfield JF, Barker WW, Welch SA, Taunton A (1999) Biological impact on mineral dissolution: application of the lichen model to understanding mineral weathering in the rhizosphere. Proc Natl Acad Sci USA 96:3404–3411
Behnam F, Vilcinskas A, Wagner M, Stoecker K (2012) A straightforward DOPE (double labeling of oligonucleotide probes)-FISH (fluorescence in situ hybridization) method for simultaneous multicolor detection of six microbial populations. Appl Environ Microbiol 78:5138–5142
Borderie F, Alaoui-Sossé B, Aleya L (2015) Heritage materials and biofouling mitigation through UV-C irradiation in show caves: state-of-the-art practices and future challenges. Environ Sci Pollut Res 22:4144–4172
Borrego S, Valdes O, Vivar I et al (2012) Essential oils of plants as biocides against microorganisms isolated from Cuban and Argentine documentary heritage. ISRN Microbiol 2012:826786
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Caneva G, Nugari MP, Salvadori O (1991) Biology in the conservation of works of art. ICCROM, Rome
Casano LM, Braga MR, Álvarez R, del Campo EM, Barreno E (2015) Differences in the cell walls and extracellular polymers of the two Trebouxia microalgae coexisting in the lichen Ramalina farinacea are consistent with their distinct capacity to immobilize extracellular Pb. Plant Sci 236:195–204
Cattò C, Cappitelli F (2019) Testing anti-biofilm polymeric surfaces: where to start? Int J Mol Sci 20:3794
Cattò C, Grazioso G, Dell’Orto SC, Gelain A, Villa S, Marzano V, Vitali A, Villa F, Cappitelli F, Forlani F (2017) The response of Escherichia coli biofilm to salicylic acid. Biofouling 33:235–251
Chamizo S, Adessi A, Mugnai G, Simiani A, De Philippis R (2018) Soil type and cyanobacteria species influence the macromolecular and chemical characteristics of the polysaccharidic matrix in induced biocrusts. Microb Ecol 78:482–493
Daddi Oubekka S, Briandet R, Fontaine-Aupart MP, Steenkeste K (2012) Correlative time-resolved fluorescence microscopy to assess antibiotic diffusion-reaction in biofilms. Antimicrob Agents Chemother 56:3349–3358
Davison WM, Pitts B, Stewart PS (2010) Spatial and temporal patterns of biocide action against Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother 54:2920–2927
De Los Ríos A, Wierzchos J, Sancho LG, Green TGA, Ascaso C (2005) Ecology of endolithic lichens colonizing granite in continental Antarctica. Lichenologist 37:383–395
Dhifi W, Bellili S, Jazi S, Bahloul N, Mnif W (2016) Essential oils’ chemical characterization and investigation of some biological activities: a critical review. Medicines (Basel) 3:25
EFSA (2012) Conclusion on the peer review of the pesticide risk assessment of the active substance thymol. EFSA J 10:2916
Ettenauer J, Piñar G, Tafer H, Sterflinger K (2015) Quantification of fungal abundance on cultural heritage using real time PCR targeting the β-actin gene. Front Microbiol 5:262
Fernandez-Silva I, Sanmartín P, Silva B, Moldes A, Prieto B (2011) Quantification of phototrophic biomass on rocks: optimization of chlorophyll-a extraction by response surface methodology. J Ind Microbiol Biotechnol 38:179–188
Franklin MJ, Chang C, Akiyama T, Bothner B (2015) New technologies for studying biofilms. Microbiol Spectrum 3. https://doi.org/10.1128/microbiolspec
Gambino M, Ahmed MAA, Villa F, Cappitelli F (2017) Zinc oxide nanoparticles hinder fungal biofilm development in an ancient Egyptian tomb. Int Biodeter Biodegr 122:92–99
Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, Mililli L, Hunt C, Lu J, Osvath SR, Monahan LG, Cavaliere R, Charles IG, Wand MP, Gee ML, Prabhakar R, Whitchurch CB (2013) Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci USA 110:11541–11546
Gorbushina AA, Broughton WJ (2009) Microbiology of the atmosphere–rock interface: how biological interactions and physical stresses modulate a sophisticated microbial ecosystem. Ann Rev Microbiol 63:431–450
Grube M, Berg G (2009) Microbial consortia of bacteria and fungi with focus on the lichen symbiosis. Fungal Biol Rev 23:72–85
Guilbaud M, de Coppet P, Bourion F, Rachman C, Prévost H, Dousset X (2005) Quantitative detection of Listeria monocytogenes in biofilms by realtime PCR. Appl Environ Microbiol 71:2190–2194
Hall CW, Mah TF (2017) Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev 41:276–301
Haussler S, Fuqua C (2013) Biofilms 2012: new discoveries and significant wrinkles in a dynamic field. J Bacteriol 195:2947–2958
Holben Ellis M (1995) The care of prints and drawings. Altamira Press, Walnut Creek
Isbell LH (1997) The effects of thymol on paper, pigments, and media. Abbey Newsletter 21:39–43
Ivanova EP, Alexeeva YV, Pham DK, Wright JP, Nicolau DV (2006) ATP level variations in heterotrophic bacteria during attachment on hydrophilic and hydrophobic surfaces. Int Microbiol 9:37–46
Jessup CM, Kassen R, Forde SE, Kerr B, Buckling A, Rainey PB, Bohannan BJM (2004) Big questions, small worlds: microbial model systems in ecology. Trends Ecol Evol 19:189–197
Jesus B, Perkins RG, Mendes CR, Brotas V, Paterson DM (2006) Chlorophyll fluorescence as a proxy for microphytobenthic biomass: Alternatives to the current methodology. Mar Biol 150:17–28
Kaplan JB (2010) Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res 89:205–218
Konkol N, McNamara C, Sembrat J, Rabinowitz M, Mitchell R (2009) Enzymatic decolorization of bacterial pigments from culturally significant marble. J Cult Herit 10:362–366
Konkol N, McNamara CJ, Mitchell R (2010) Fluorometric detection and estimation of fungal biomass on cultural heritage materials. J Microbiol Methods 80:178–182
Kuhn DM, Balkis M, Chandra J, Mukherjee PK, Ghannoum MA (2003) Uses and limitations of the XTT assay in studies of Candida growth and metabolism. J Clin Microbiol 41:506–508
Kyi C, Sloggett R, Schiesser CH (2014) Preliminary investigations into the action of nitric oxide in the control of biodeterioration. AICCM Bull 35:60–68
López AJ, Rivas T, Lamas J, Ramil A, Yáñez A (2010) Optimisation of laser removal of biological crusts in granites. Appl Phys A-Mater Sci Process 100:733–739
Macro N, Sbrana C, Legnaioli S., Galli E (2018) Antimicrobial activity of essential oils: a green alternative to treat cultural heritage. In: Mosquera MJ, Almoraima Gil ML (eds) Conserving cultural heritage. Proceedings of the 3rd International Congress on Science and Technology for the (TechnoHeritage 2017), May 21–24, 2017, Cadiz, Spain, pp 291–293
Martin-Sanchez PM, Nováková A, Bastian F, Alabouvette C, Saiz-Jimenez C (2012) Use of biocides for the control of fungal outbreaks in subterranean environments: the case of the Lascaux Cave in France. Environ Sci Technol 46:3762–3770
Merritt JH, Kadouri DE, O’Toole GA (2005) Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1, Unit–1B.1, 1B.1.1–1B.1.17
Miller AZ, Laiz L, Gonzalez JM, Dionísio A, Macedo MF, Saiz-Jimenez C (2008) Reproducing stone monument photosynthetic–based colonization under laboratory conditions. Sci Total Environ 405:278–285
Miller AZ, Liaz L, Dionisio A, Macedo MF, Saiz-Jimenez C (2009) Growth of phototrophic biofilms from limestone monuments under laboratory conditions. Int Biodeter Biodegr 63:860–867
Nante N, Ceriale E, Messina G, Lenzi D, Manzi P (2017) Effectiveness of ATP bioluminescence to assess hospital cleaning: a review. J Prev Med Hyg 58:E177–E183
Neu TR, Lawrence JR (2014) Investigation of microbial biofilm structure by laser scanning microscopy. Adv Biochem Eng Biotechnol 146:1–51
Nezhadali A, Nabavi M, Rajabian M, Akbarpour M, Pourali P, Amini F (2014) Chemical variation of leaf essential oil at different stages of plant growth and in vitro antibacterial activity of Thymus vulgaris Lamiaceae, from Iran. Beni-Suef Univ J Basic Appl Sci 3:87–92
Noack-Schönmann S, Bus T, Banasiak R, Knabe N, Broughton WJ, Dulk-Ras HD, Hooykaas PJJ, Gorbushina AA (2014) Genetic transformation of Knufia petricola A95 – a model organism for biofilm–material interactions. AMB Express 4:80
Olson ME, Ceri H, Morck DW, Buret AG, Read RR (2002) Read biofilm bacteria: formation and comparative susceptibility to antibiotics. Can J Vet Res 66:86–92
Pantanella F, Valenti P, Natalizi T, Passeri D, Berlutti F (2013) Analytical techniques to study microbial biofilm on abiotic surfaces: pros and cons of the main techniques currently in use. Ann Ig 25:31–42
Piñar G, Sterflinger K, Ettenaueret J, Quandt A, Pinzari F (2015) A combined approach to assess the microbial contamination of the archimedes palimpsest. Microb Ecol 69:118–134
Polo A, Diamanti MV, Bjarnsholt T, Høiby N, Villa F, Pedeferri MP, Cappitelli F (2011) Effects of photoactivated titanium dioxide nanopowders and coating on planktonic and biofilm growth of Pseudomonas aeruginosa. Photochem Photobiol 87:1387–1394
Rakotonirainy MS, Lavédrine B (2005) Screening for antifungal activity of essential oils and related compounds to control the biocontamination in libraries and archives storage areas. Int Biodeter Biodegr 55:141–147
Robertson PKJ, Alakomi HL, Arrien N, Gorbushina AA, Krumbein WE, Maxwell I, McCullagh C. Ross N, Saarela M, Valero J, Vendrell M, Young ME (2004) Inhibitors of biofilm damage on mineral materials (Biodam). In: Kwiatkowski D, Löfvendahl R (eds) 10th International Congress on Deterioration and Conservation of Stone, Stockholm, Sweden, 27 June-2 July 2004. ICOMOS Sweden, pp 399–406
Rosado T, Martins MR, Pires M, Mirão J, Candeias A, Caldeira AT (2013) Enzymatic monitorization of mural paintings biodegradation and biodeterioration. Int J Conserv Sci 4:603–612
Ruggiero L, Bartoli F, Fidanza MR, Zurlo F, Marconi E, Gasperi T, Tuti S, Crociani L, Di Bartolomeo E, Caneva G, Ricci MA, Sodo A (2020) Encapsulation of environmentally-friendly biocides in silica nanosystems for multifunctional coatings. Appl Surf Sci 514:145908
Sanmartín P, Aira N, Devesa-Rey R, Silva B, Prieto B (2010) Relationship between color and pigment production in two stone biofilm-forming cyanobacteria (Nostoc sp PCC 9104 and Nostoc sp PCC 9025). Biofouling 26:499–509
Sanmartín P, Villa F, Polo A, Silva B, Prieto B, Cappitelli F (2015) Rapid evaluation of three biocide treatments against the cyanobacterium Nostoc sp. PCC 9104 by color changes. Ann Microbiol 65:1153–1158
Sebestyen P, Blanken W, Bacsa I, Tóth G, Martin A, Bhaiji T, Dergez Á, Kesseru P, Koós Á, Kiss I (2016) Upscale of a laboratory rotating disk biofilm reactor and evaluation of its performance over a half-year operation period in outdoor conditions. Algal Res 18:266–272
Seiffert F, Bandow N, Bouchez J, von Blanckenburg F (2014) Microbial colonization of bare rocks: laboratory biofilm enhances mineral weathering. Procedia Earth Planet Sci 10:123–129
Seiffert F, Bandow N, Kalbe U, Milke R, Gorbushina AA (2016) Laboratory tools to quantify biogenic dissolution of rocks and minerals: a model rock biofilm growing in percolation columns. Front Earth Sci 4:31
Sendersky E, Simkovsky R, Golden SS, Schwarz R (2017) Quantification of chlorophyll as a proxy for biofilm formation in the cyanobacterium Synechococcus elongatus. Bio-Protoc 7:14
Shadia MAA, Aeron A (2014) Bacterial biofilm: dispersal and inhibition strategies. SAJ Biotechnol 1:105
Silva LN, Zimmer KR, Macedo AJ, Silva Trentin D (2016a) Plant natural products targeting bacterial virulence factors. Chem Rev 116:9162–9236
Silva M, Salvador C, Candeias MF, Teixeira D (2016b) Toxicological assessment of novel green biocides for cultural heritage. Int J Conserv Sci 7:265–272
Spadoni Andreani E, Villa F, Cappitelli F, Krasowska A, Biniarz P, Łukaszewicz M, Secundo F (2017) Coating polypropylene surfaces with protease weakens the adhesion and increases the dispersion of Candida albicans cells. Biotechnol Lett 39:423–428
Spiers AJ, Bohannon J, Gehrig SM, Rainey PB (2003) Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol Microbiol 50:15–27
Urzì C, De Leo F, Krakova L, Pangallo D, Bruno L (2016) Effects of biocide treatments on the biofilm community in Domitilla’s catacombs in Rome. Sci Total Environ 572:252–262
Valentini F, Diamanti A, Palleschi G (2010) New bio-cleaning strategies on porous building materials affected by biodeterioration event. Appl Surf Sci 256:6550–6563
Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG (2011) Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci USA 108:4152–4157
Valm AM, Mark Welch JL, Borisy GG (2012) CLASI-FISH: principles of combinatorial labeling and spectral imaging. Syst Appl Microbiol 35:496–502
Vázquez-Nion D, Rodríguez-Castro J, López-Rodríguez MC, Fernández-Silva I, Prieto B (2016) Subaerial biofilms on granitic historic buildings: microbial diversity and development of phototrophic multi–species cultures. Biofouling 32:657–669
Vazquez-Nion D, Silva B, Prieto B (2018) Bioreceptivity index for granitic rocks used as construction material. Sci Total Environ 633:112–121
Villa F, Remelli W, Forlani F, Vitali A, Cappitelli F (2012) Altered expression level of Escherichia coli proteins in response to treatment with the antifouling agent zosteric acid sodium salt. Environ Microbiol 14:1753–1761
Villa F, Pitts B, Lauchnor E, Cappitelli F, Stewart PS (2015) Development of a laboratory model of a phototroph–heterotroph mixed–species biofilm at the stone/air interface. Front Microbiol 6:1251
Villa F, Gulotta D, Toniolo L, Borruso L, Cattò C, Cappitelli F (2020) Aesthetic alteration of marble surfaces caused by biofilm formation: effects of chemical cleaning. Coatings 10:122
Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R (2013) Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168
Vujcic I, Masic S, Medic M (2019) The influence of gamma irradiation on the color change of wool, linen, silk, and cotton fabrics used in cultural heritage artifacts. Radiat Phys Chem 156:307–313
Welch K, Cai Y, Strømme M (2012) A method for quantitative determination of biofilm viability. J Funct Biomater 3:418–431
Wu J, Lou W (2016) Study of biological enzymes’ inhibition of filamentous fungi on the Crosslake Bridge ruins. Sci Archaeol Conserv:25–29
Xie Z, Thompson A, Kashleva H, Dongari-Bagtzoglou A (2011) A quantitative real-time RT-PCR assay for mature C. albicans biofilms. BMC Microbiol 11:93
Young ME, Alakomi H-L, Fortune I, Gorbushina AA, Krumbein WE, Maxwell I, McCullagh C, Robertson P, Saarela M, Valero J, Vendrell M (2008) Development of a biocidal treatment regime to inhibit biological growths on cultural heritage: BIODAM. Environ Geol 56:631–641
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this chapter
Cite this chapter
Cappitelli, F., Villa, F. (2021). Novel Antibiofilm Non-Biocidal Strategies. In: Joseph, E. (eds) Microorganisms in the Deterioration and Preservation of Cultural Heritage. Springer, Cham. https://doi.org/10.1007/978-3-030-69411-1_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-69411-1_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-69410-4
Online ISBN: 978-3-030-69411-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)