Abstract
The evolution and structure of prokaryotic genomes are largely shaped by horizontal gene transfer. This process is so prevalent that DNA can be seen as a public good—a resource that is shared across individuals, populations, and species. The consequence is a network of DNA sharing across prokaryotic life, whose extent is becoming apparent with increased availability of genomic data. Within prokaryotic species, gene gain (via horizontal gene transfer) and gene loss results in pangenomes, the complete set of genes that make up a species. Pangenomes include core genes present in all genomes, and accessory genes whose presence varies across strains. In this chapter, we discuss how we can understand pangenomes from a network perspective under the view of DNA as a public good, how pangenomes are maintained in terms of drift and selection, and how they may differ between prokaryotic groups. We argue that niche adaptation has a major impact on pangenome structure. We also discuss interactions between accessory genes within genomes, and introduce the concepts of ‘keystone genes’, whose loss leads to concurrent loss of other genes, and ‘event horizon genes’, whose acquisition may lead to adaptation to novel niches and towards a separate, irreversible evolutionary path.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Horizontal Gene Transfer (HGT) is the most important force affecting evolutionary change in prokaryotes, and its pervasiveness has resulted in a vast global network of connectivity between microorganisms. DNA is available for horizontal acquisition by prokaryotes in a variety of ways: conjugative plasmids (Grohmann et al. 2003; Lederberg and Tatum 1946) facilitate the transfer of DNA directly from cell to cell, phage can facilitate the indirect movement of DNA from one prokaryotic cell to another by generalised transduction (Zinder and Lederberg 1952), and gene transfer agents (GTAs) facilitate gene transfer by cell lysis. In some Archaea, we even see the formation of networks of connections between individuals that can lead to the formation of heterodiploid cells and recombination between the parental cells’ genomes (Naor and Gophna 2013). Another important mechanism is direct acquisition of DNA through transformation. Extracellular DNA has a ubiquitous distribution in natural environments from hydrothermal vents, to freshwater, soil, and sediment (Nagler et al. 2018), as well as in the biofilms (Steinberger and Holden 2005) that line our sewage pipes (Vincke et al. 2001), contaminate hospital equipment (Stickler 2008), associate with tooth decay (Potera 1999), and much more. Therefore, DNA can be shared and used among organisms and effectively becomes a public good. All these mechanisms result in a DNA-sharing network that has probably existed since before life evolved to become cellular and will likely remain an important part of prokaryotic biology for as long as there are prokaryotes.
With the advent, and subsequent accessibility, of next-generation sequencing technologies (Shendure et al. 2017), it became apparent that gene presence–absence variability within a species (i.e. strain-to-strain variability) was much larger than expected (Tettelin et al. 2005). For example, when the first three Escherichia coli genomes were sequenced, only 39.2% of their protein-coding genes were found to be common to all three genomes (Welch et al. 2002). In a more recent study involving 1524 Pseudomonas aeruginosa genomes, only 3% of genes were found to be shared (i.e. ‘core’) across all strains, with the remaining 97% being variably present in a subset of strains (Karasov et al. 2018). The existence of this variability in gene content within what we regard as single prokaryotic species led to the concept of a pangenome, the complete set of genes that are present in a given species (Tettelin et al. 2005). This set of genes is usually divided into two categories: core genes, that are present across all individuals in a species, and accessory genes, whose presence varies between individuals or strains (Tettelin et al. 2005; Welch et al. 2002; Karasov et al. 2018; Laing et al. 2010). The pangenome concept revolutionises our thinking, since it means considering organisms like Escherichia coli not only in terms of the thousand or so genes that are common to all members, but also in terms of the 100,000 or so genes that are found in at least one, but not all, E. coli genomes (Land et al. 2015). This new information on the structure of the prokaryotic world has meant that we have to think about ‘units’ of selection (Okasha 2006) in different ways. In this chapter, we will outline some of the ways in which we can think about pangenomes and what this means for biology. Although our focus is on prokaryotes, it should be noted that some eukaryotes also have pangenomes. For example, a high degree of gene presence–absence polymorphism has been found in different genome sequences of humans (Sherman et al. 2019), cultivated rice (Wang et al. 2018; Hubner et al. 2019), sunflower (Hubner et al. 2019), and in the widespread coccolithophore Emiliania huxleyi (Read et al. 2013).
2 Pangenome Properties
As a consequence of the merging of genetic information through HGT and the existence of pangenomes, our thinking about the evolutionary history of prokaryotic genomes has changed. In fact, it is more relevant to think not of the evolutionary history of a genome, but rather the evolutionary histories of the various parts of a genome, since these histories can be different (Bapteste et al. 2009). The phylogenetic relationships inferred by a single gene, no matter how important that gene, rarely reflects the evolutionary history of the suite of organisms under consideration. This idea was codified by Darwin in ‘The Origin’ when he said: ‘The importance, for classification, of trifling characters, mainly depends on their being correlated with several other characters of more or less importance’ (Darwin 1860). In other words, the notion of homoplastic characters (i.e. characters whose similarity is due to convergent evolution) is an old idea and characters can differ in what they suggest is the proper classification of an organism. Though Darwin did not know about DNA or HGT, the warning about character congruence and classification still holds true today and perhaps even more so because of HGT and the non-tree likeness of this process.
The pangenomes of different prokaryotic groups differ. Transformation, transduction, and conjugation contribute to shuffling variably sized portions of genomes through both homologous and non-homologous recombination. The frequency of the different mechanisms likely depends on environmental conditions, lifestyle, and cell biology (i.e. the molecular mechanisms present in particular cells or taxa) (Hanage 2016). Therefore, under different conditions, HGT and recombination can in principle range from non-existent to widespread, resulting in primarily clonal or panmictic groups, respectively (Yang et al. 2019). Furthermore, recombination barriers, both within and between species, can be fuzzy and potentially differ for different parts of the genome. This can make the delineation of populations or of species more complicated in prokaryotes, when compared to animals, for example (Hanage 2013). However, it has been suggested that natural species boundaries do exist in prokaryotes and that they can be defined (Bobay and Ochman 2017). On the whole, HGT and DNA recombination in prokaryotes can have similar consequences to sexual reproduction in eukaryotes: removing deleterious mutations, thereby avoiding Muller’s ratchet or mutational meltdown, while also offering a mechanism for bringing together advantageous mutations in different genes or parts of the genome. But crucially in prokaryotes, recombination can both remove and add a hugely variable number of genes to a genome, thereby affecting the overall gene repertoire rather than simply modifying existing genes by point mutation. That is, recombination in prokaryotes often results in insertions or deletions, while in eukaryotes it tends to swap alleles between chromosomes.
Pangenomes differ in the degree to which they are ‘open’ or ‘closed’. Species that share almost all genes with each other (i.e. have very little strain-to-strain gene content dissimilarity) having a large ‘core’ and small ‘accessory’ genome, are considered to have closed pangenomes (McInerney et al. 2017). In contrast, species can have open pangenomes in which gene content varies appreciably from one genome to another (McInerney et al. 2017) (see Fig. 1). Though we know the openness of prokaryotic pangenomes varies greatly from one species to the next (Tettelin et al. 2005), our estimates of openness can be affected by the available genomic data (i.e. the number of accessory genes is expected to increase as more strain information becomes available). As such, openness can be measured by modelling the number of accessory genes as a function of the number of sequenced genomes (Tettelin et al. 2005) (see also Chap. 1). The first analysis of openness found that eight Streptococcus agalactiae genomes were not enough to uncover all possible accessory genes and predicted that new genes would be found with every additional genome, leading to an essentially infinite pangenome. In contrast, the number of new accessory genes in Bacillus anthracis dropped to zero after the incorporation of only four genomes to the study of its pangenome (Tettelin et al. 2005). Therefore, accurate measurements of pangenome openness depend on sampling the broad diversity of genomes in a given species, and such measurements should ideally account for core genome diversity and the phylogenetic relationships between those genomes.
3 Public Goods
The idea that DNA functions as a public good (Erwin 2015; McInerney et al. 2011a; McInerney and Erwin 2017) stems from the fact that HGT makes DNA available to other ‘users’ and this process has structured a great deal of the life on this planet, both cellular and viral (Bapteste et al. 2012, 2013). Integration of a new DNA sequence into a genome can only be successful if the source organism and the recipient organism can both make use of this DNA in some way. Carl Woese referred to the universal genetic code as being the ‘lingua franca’ of genetic commerce (Woese 2002). HGT has been observed in almost all known phyla, though HGT seems to be reduced in frequency among eukaryotes and perhaps reduced further in multicellular organisms (Schonknecht et al. 2014; McInerney et al. 2014; Ku et al. 2015). As a consequence of HGT, there is no universal Tree of Life, and instead there is a network of life reflecting the vertical and horizontal movements of genetic information (Bapteste et al. 2012, 2013; Corel et al. 2018).
Our current appreciation of evolutionary history in prokaryotes and the observations of pangenomes has led us to consider what metaphors might be appropriate for representing, modelling, and understanding life on the planet. A variety of alternatives to the tree metaphor, such as ‘cobwebs of life’ (Ge et al. 2005) or ‘rhizome of life’ (Merhej et al. 2011), have been used. However, some of us have proposed to depart from a way of thinking that inherently depends on a particular kind of diagram. Instead we have advocated a focus on the fundamental process of HGT, and the fact that it constructs new genomes in the same way that, say, a furniture manufacturing plant might bring together different materials in order to construct a new kind of chair, or in the way that a football team might substitute one player for another. As mentioned above, Woese suggested that HGT could be thought of in commercial terms (Woese 2002), and a logical extension to this line of thinking is that DNA acts as though it is a ‘public good’ (McInerney et al. 2011a, b; McInerney and Erwin 2017). Briefly, in the theory of goods, Nobel laureate Paul Samuelson initially described two kinds of goods thus: ‘[…] I explicitly assume two categories of goods: ordinary private consumption goods which can be parcelled out among different individuals […] and collective consumption goods […] which all enjoy in common in the sense that each individual’s consumption of such a good leads to no subtraction from any other individual’s consumption of that good […]’ (Samuelson 1954). Since then, the concept has been expanded so that four kinds of goods are recognised—private goods, public goods, club goods, and common goods (McInerney et al. 2011a), based on whether goods are rivalrous and/or excludable. The criteria for each of the classifications are contained in Fig. 2, along with examples of goods that fall easily into each of these categories. A ‘good’ is said to be rivalrous if its consumption by one consumer prevents simultaneous consumption by other consumers, and a ‘good’ is said to be excludable if it is possible to prevent others from having access to it. DNA possesses the property of being non-excludable (e.g. the DNA of any individual is made available, at least at the time of death of the cell or the individual) and it is also non-rivalrous in a practical sense, given that the amount of DNA that is produced by any given species cannot realistically be used up by any consumer. This perspective is useful in the sense that viewing genome evolution as a process of building functioning tools (i.e. new kinds of organisms) allows us to ask questions that would not make much sense if we used ‘tree-thinking’ (Bapteste et al. 2013; Dagan and Martin 2009). Tree-thinking inherently supposes that genes came to be in a genome because all the genes have been inherited through the same lineage of descent—a process that infers that genes are ‘private’ to a clade. ‘Goods-thinking’, on the other hand, frees us to think more about why the particular set of genes that we observe in a genome are there, rather than some other set of genes. We do not assume that any gene is a private good, exclusively found in a particular species or clade, with other organisms excluded from accessing the segment of DNA. Goods-thinking infers that a genome has evolved to be the way it is through vertical inheritance from a common ancestor, but also through the horizontal acquisition of genes, with the rate of gain (and loss) of genes being modified by the influences of random drift, selection, and demography. Goods-thinking, therefore, needs some new tools, outside of the framework of the bifurcating phylogenetic tree, in order to properly analyse gene and genome evolution (Bapteste et al. 2009). Here we deal specifically with the pangenome’s part of Goods Thinking theory.
4 Analyses of Pangenomes
Because of the fluidity of genomes, caused by accessory gene gain and loss, the analysis of pangenomes lends itself more suitably to networks than to phylogenies. Networks are mathematical graphs represented by nodes, or vertices, which are connected by edges, or lines, if-and-only-if a relationship exists between them. Networks are widely used in ecology—and in biology in general—to represent, for example food webs (Dunne et al. 2002), social interactions (Robins et al. 2007), nutrient/energy flows (Allesina et al. 2005), and cooperation between members in a population (Jain and Krishna 2001). Networks can have edges that are either directed (often shown as an arrow) or undirected, depending on whether the relationship that connects the nodes has directionality (e.g. to connect an organism to their food source in a food web). The study of networks, or graphs (i.e. graph theory) dates to at least 1735 (Skiena 2008; Compeau et al. 2011) and has advanced rapidly due to its applications in computer science, engineering, physics, and biology. The public goods nature of DNA makes a network structure ideal to uncover patterns and processes of evolution in ways where phylogenetic trees would be somewhat lacking, since phylogenetic trees do not infer lateral movement of genetic material. The analysis of features contained within the graphs such as non-transitive triplets, or nodes with identical incident edges can reveal patterns of recombination or gene sharing (Bapteste et al. 2012; Corel et al. 2018; Meheust et al. 2018).
In the analysis of pangenomes, networks are often k-partite or multi-partite, meaning that their nodes can be coloured using k colours such that no node is directly connected to another with the same colour (Pavlopoulos et al. 2018). A special case of k-partite graphs is bipartite or two colourable graphs. In pangenome analyses, bipartite graphs usually connect genomes to their constituent genes (Corel et al. 2018). Bipartite networks have been used previously to identify the levels of gene sharing within microbial genomes (Corel et al. 2018), to characterise the capacity of accessory genes in metabolic networks (Goyal 2018), and to interrogate gene presence/absence patterns and coincident relationships (McNally et al. 2016).
Especially relevant for genome evolution is the N-rooted fusion graph (Haggerty et al. 2014). This graph differs from a phylogenetic tree due to the presence of more than one root node (a node that depicts the point-of-origin of all operational taxonomic units in the graph) and the presence of at least one internal node in the graph where the in-degree of the node (the number of edges pointing towards that node) is greater than 1 and the out-degree of the node (the number of edges emerging from that node) is 1 (Fig. 3). In other words, the merging of genetic material inherently means that the graph needs more than a single origin or root. It also means that the point at which the material merged must be represented by a merger, or fusion node (Fig. 3). The various components of the internal structure of an N-rooted fusion graph can be determined by the usual phylogenetic approaches [i.e. parsimony, likelihood, or distance matrix methods (Felsenstein 2003)]. The complete N-rooted graph is then constructed by merging of these individual phylogenetic trees, by constructing fusion nodes at the appropriate places (Haggerty et al. 2014).
An N-Rooted Fusion Graph. This kind of branching diagram can be used to illustrate the merging of evolving objects. The nodes labelled R indicate the root nodes for this graph. Each root node depicts the root for a different kind of gene. The node labelled F indicated the fusion node. The different node colours indicate different gene families, with the blue nodes indicating that they are a fusion family
5 How Are Pangenomes Maintained?
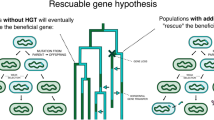
Because acquired DNA can function across multiple organisms—facilitating it to become a public good—HGT into some individuals in a population creates diversity within that species. Transferred sequences will be present in a subset of the population’s genomes and absent in the rest (McNally et al. 2016), becoming raw material for natural selection (see Fig. 4). Multiple iterations of this process have most likely resulted in the observed pattern of hugely varying gene content across conspecific genomes (Welch et al. 2002; Lukjancenko et al. 2010; Koonin and Wolf 2008). Maintenance of the observed high levels of variation requires an explanation, because, while we know that transformation, conjugation, and transduction introduce this presence–absence variation, it is expected that both natural selection and genetic drift would remove this kind of genetic variation from populations. In terms of sequence variation within populations, different mechanisms have been proposed to explain the maintenance of diversity. These mechanisms range from relatively trivial explanations, such as the existence of a balance between the rates at which new variants arise in populations (by mutation, for example) and the rates at which they are removed, to more exotic mechanisms such as heterozygote advantage, interactions between genotypes and different environments, and negative frequency-dependent selection (Hahn 2018). Although most of these explanations have been developed in order to account for high levels of genetic diversity in diploid, sexually reproducing eukaryotes, some of these mechanisms can also help us to understand genetic variation in prokaryotes. However, understanding the existence and maintenance of pangenomes has its own particular challenges.
A key element to be considered when we speak about mechanisms that maintain variability in gene content in prokaryotic populations is the fitness effect that these accessory genes have on individuals. We will likely find examples of particular genes whose presence is neutral, deleterious, or adaptive in most genomes; we are already familiar with genes in the latter class such as those conferring antibiotic resistance and pathogenicity islands (Sheppard et al. 2018). However, an interesting question to think about is whether accessory genes on average contribute to fitness (or under which circumstances they may be adaptive), and which mechanisms have led to their patchy occurrence in genomes. Depending on the average fitness effect of accessory genes, different mechanisms could be governing their presence.
If accessory genes are mostly deleterious, which could be the case if they are predominantly selfish or parasitic, then the patchy presence patterns that we observe could reflect a constant arms race between these selfish elements and the host genome (somewhat equivalent to the Red Queen hypothesis for maintenance of variability in populations of interacting hosts and pathogens). Although this pattern may be responsible for a proportion of accessory genes, it is very unlikely that this explains most of the observed variability and the existence of pangenomes, partly because many accessory genes are not related to selfish elements and appear to be involved in multiple cellular functions (McNally et al. 2016; Sheppard et al. 2018).
If accessory genes are usually neutral in terms of fitness, eventually they would be randomly fixed or lost in different populations due to genetic drift, particularly if recombination is rare. A neutralist model for pangenomes implies that we see presence–absence variation because there is a random ‘rain’ of genes constantly being acquired and we observe their presence in a genome because they have either not had enough time to drift to fixation or to be lost again. This kind of model implies that neither the gain nor the loss of accessory genes has a fitness effect (Baumdicker et al. 2012), a situation that seems contradicted by the observation of both prophage (Nanda et al. 2015) and antibiotic resistance (Her and Wu 2018) genes affecting fitness. A recent study (Andreani et al. 2017) showed a correlation between pangenome fluidity and synonymous variation, which was taken to imply that genome content diversity is mostly neutral. The implication was that synonymous diversity arises in the absence of selection and if this correlates with genome fluidity, then genome fluidity is also neutral. The problem with this model is that synonymous diversity in prokaryotes is not necessarily neutral, and we see stronger selection on synonymous codon usage in organisms with large effective population size (N e) (Sharp et al. 1993), so the correlation between large N e and genome fluidity is unlikely to be a consequence of drift alone.
Recently, a drift-barrier model for pangenome evolution has been proposed (Bobay and Ochman 2018). The authors observed a positive correlation between pangenome size and N e (using two independent measures of N e for different bacterial species). In contrast to Andreani et al. (2017) they propose that, on average, accessory genes make a positive contribution to fitness. Based on nearly neutral evolutionary theory, they then explain the correlation between N e and pangenome size by the loss of slightly advantageous genes in populations with small N e. Therefore, populations with large N e would maintain a larger number of accessory genes. However, while this may help explain larger genome size (i.e. the maintenance of more genes), it does not necessarily explain diversity in gene content in different individuals from the same population, since those slightly advantageous genes would be expected to eventually fix in the population. Furthermore, the authors did not deal with the likelihood that, on occasion, these advantageous genes would result in sweeps to fixation. The problem with this model is outlined in simulations by Niehus et al. (2015). As some of us have previously proposed (McInerney et al. 2017), some of the basics of this drift-barrier model, if combined with niche adaptation, can go further in explaining the maintenance of genome content diversity. Under the adaptive pangenomes model of McInerney et al. (2017), accessory genes make, on average, a positive contribution to fitness, and this contribution may be niche dependent. Therefore, genes are maintained in the niches where they are beneficial and lost in others. However, ongoing migration would still allow recombination in other parts of the genome, and thus maintenance of large N e, at least for the core genome.
In line with the McInerney et al. (2017) model of pangenome maintenance by a combination of drift and niche-dependence, there is evidence that at least a significant fraction of accessory genes are beneficial and involved in niche adaptation (Bruns et al. 2018; Rubino et al. 2017; McInerney 2013). The adaptability of prokaryotes means that they occupy niches all over the planet—including oceans (Sunagawa et al. 2015), ice sheets (Anesio et al. 2017), and salt flats (Caton et al. 2004), as well as ecosystems deep within the earth’s crust (Chivian et al. 2008), and on and within our own bodies (The Human Microbiome Project Consortium 2012). Some ‘specialist’ prokaryotic species focus on one, specific niche; for example Buchnera aphidicola is an endosymbiont that forms an obligate association with aphids (van Ham et al. 2003). Such specialists would likely have little to gain from extensive gene content diversity, possibly explaining the relative closeness of some species pangenomes. For example, Tropheryma whipplei, an intracellular human pathogen and the causative agent of Whipple’s disease (Gorvel et al. 2010), has an extremely restricted pangenome (Fenollar et al. 2014), and smaller than average N e (Bobay and Ochman 2018). In contrast, ‘generalist’ prokaryotic species can occupy many of the niches made available to them. Escherichia coli has been identified in several different kinds of environments including the gut and urinary tract of humans, and indeed other warm- and cold-blooded animals (Tenaillon et al. 2010), as well as soil, sediment, and water (Savageau 1983). In order to occupy such variable environments, these species must be able to adapt to different carbon and nitrogen sources (Bertin et al. 2011), to evade various antibiotic pressures (Sáenz et al. 2004), and to utilise different types of respiration depending on oxygen availability (Jones et al. 2007). Recent work on the metabolic potential of accessory genes has identified a correlation between the number of novel metabolites that a given strain can synthesise and the openness of their pangenome, suggesting that the acquisition of such genes is adaptive (Goyal 2018). Other scenarios where variation in accessory genes is actively maintained by selection include negative frequency-dependent selection (Corander et al. 2017) where a major allele (gene presence or absence in our case) is at a disadvantage compared with the minor allele (the other character state). For example, in the case of vaccine programmes, it is likely that a vaccine targeting a non-essential accessory gene will confer a selective advantage on strains that do not have that accessory gene (Azarian et al. 2018). Bacteriophages may have a similar effect on non-essential attachment proteins and other cellular components. Finally, it is also the case that a particular gene may be beneficial in a specific niche when another gene is present, but not so when that partner is absent. This co-dependency of genes for fitness/adaptation to a particular niche will manifest particular patterns of co-occurrence in genomes (Cohen et al. 2013).
Notwithstanding the argument being made here that pangenomes are, on average, constructed and maintained by niche adaptation, we are still a long way from having enough data to say that this understanding is true in all cases. To assess whether neutralist or selectionist scenarios warrant greater or lesser support in different prokaryotic species and populations, we need more genomic data and information on population structure, levels of migration and recombination, and the distribution of fitness effects of accessory genes in different niches or environments. This requires deep sampling of prokaryotic genomes across space (within and between niches) and ideally along time. Recording of information on as many environmental variables as possible would also be highly advantageous for understanding which factors influence the evolution of pangenomes.
6 Keystone Genes and Event Horizon Genes
The dynamics of accessory gene repertoires is clearly a subject of great interest in microbiology. We have a poor understanding of how these repertoires are structured and what influences their content, how they grow and are maintained. The process of gene loss is also poorly understood. We have outstanding questions about what we might term ‘keystone genes’, those genes that play a central role in determining what other genes might be successful in a genome. This keystone gene concept is analogous to the keystone species concept in macroecology (Paine 1969); keystone species are those whose presence or absence can result in a major shift in the make-up of a particular ecosystem, often resulting in ecosystem collapse, if the keystone species leaves or goes extinct (Estes et al. 1978).
In a related, but slightly different context, we might consider the case of ‘event horizon’ genes. To give an example of the possible existence of such genes, we can consider the evolution by gene acquisition of Archaeal halophiles from an ancestor that was a methanogen (Nelson-Sathi et al. 2012). This transition must have involved the rapid acquisition of a large number of genes. Whereas Haloarchaea are heterotrophic, facultatively anaerobic or aerobic organisms with a phototrophic capability, their ancestors the methanogens are obligately anaerobic, methane-producing, chemolithotrophic archaea. The differences between these two closely related groups illustrate that seismic changes in genome content can occur, but also that the absence of intermediate forms suggests that such changes can come about with great rapidity. This leads us to the question of which genes, when acquired, led to the establishment of the halophile phenotype. In an analogy with astrophysics, we can speculate whether there has been an ‘event horizon’ or a point of no return, where the acquisition of a particular gene or set of genes permanently converted a methanogen to a halophile. We might imagine that the combination of importers of organic compounds and genes for heterotrophic metabolism marked the point of no return. Indeed, there seems to have been in this case no return, since all halophilic archaea are monophyletic and none have abandoned this lifestyle. Therefore, the order of gene acquisition and gene loss is an important question. Future work will help understand whether these keystone and event horizon genes are common in accessory gene repertoires.
7 Some Conclusions and Future Directions
While evolution has no particular direction, the likely success of a particular genomic sequence relates to the notion of ‘unity of purpose’. In this sense, the various components of a biochemical pathway can be said to have unity of purpose—collectively they enable the biological transformation of some important molecules. The components of the translation apparatus similarly have a unity of purpose. As a corollary, we could say that inserting genes that can enable methanogenesis into the same genome as genes that are responsible for importing sugars would not likely lead to a genome with a particularly united purpose—one part of the genome would be dedicated to producing energy by chemolithotrophy, while another part of the genome would be dedicated to a heterotrophic lifestyle. Yet situations like this must surely arise from time to time, given the pervasiveness of HGT. Two great unknowns right now include how often such conflicts arise in nature, and how compatible are the genes we see in genomes. We know that they are compatible enough to give rise to functioning organisms, but we do not know how each individual gene contributes to fitness. Background selection and hitch-hiking Hill-Robertson effects (Hill and Robertson 1966) are mechanisms that can limit the ‘impact’ of natural selection and allow maintenance of slightly deleterious variants (Price and Arkin 2015), including, we would suppose, accessory genes that have a slightly deleterious fitness effect.
The focus on pangenomes is usually centred on protein-coding genes, but there are several other levels at which pangenomes provide food for thought. An analysis of E. coli genomes has revealed that selection on non-coding regions has been instrumental in shaping the success of a particular sequence type (ST131) of the species (McNally et al. 2016). This brings into focus the combinatorial nature of genome structure—that the presence or absence of particular kinds of protein-coding genes, or even RNA-coding genes is only part of the story, and that the ‘regulatory pangenome’ will be one of the most important future challenges.
References
Allesina S, Bodini A, Bondavalli C (2005) Ecological subsystems via graph theory: the role of strongly connected components. Oikos 110(1):164–176
Andreani NA, Hesse E, Vos M (2017) Prokaryote genome fluidity is dependent on effective population size. ISME J 11(7):1719–1721
Anesio AM et al (2017) The microbiome of glaciers and ice sheets. NPJ Biofilms Microbiomes 3:10
Azarian T et al (2018) Prediction of post-vaccine population structure of Streptococcus pneumoniae using accessory gene frequencies. bioRxiv. https://doi.org/10.1101/420315
Bapteste E et al (2009) Prokaryotic evolution and the tree of life are two different things. Biol Direct 4(1):34
Bapteste E et al (2012) Evolutionary analyses of non-genealogical bonds produced by introgressive descent. Proc Natl Acad Sci USA 109(45):18266–18272
Bapteste E et al (2013) Networks: expanding evolutionary thinking. Trends Genet 29(8):439–441
Baumdicker F, Hess WR, Pfaffelhuber P (2012) The infinitely many genes model for the distributed genome of bacteria. Genome Biol Evol 4(4):443–456
Bertin Y et al (2011) Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol 13(2):365–377
Bobay LM, Ochman H (2017) Biological species are universal across life’s domains. Genome Biol Evol 9(3):491–501
Bobay L-M, Ochman H (2018) Factors driving effective population size and pan-genome evolution in bacteria. BMC Evol Biol 18(1):153
Bruns H et al (2018) Function-related replacement of bacterial siderophore pathways. ISME J 12(2):320–329
Caton TM et al (2004) Halotolerant aerobic heterotrophic bacteria from the Great Salt Plains of Oklahoma. Microb Ecol 48(4):449–462
Chivian D et al (2008) Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322(5899):275–278
Cohen O et al (2013) CoPAP: coevolution of presence-absence patterns. Nucleic Acids Res 41(Web Server issue):W232–W237
Compeau PEC, Pevzner PA, Tesler G (2011) Why are de Bruijn graphs useful for genome assembly? Nat Biotechnol 29(11):987
Corander J et al (2017) Frequency-dependent selection in vaccine-associated pneumococcal population dynamics. Nat Ecol Evol 1(12):1950–1960
Corel E et al (2018) Bipartite network analysis of gene sharings in the microbial world. Mol Biol Evol 35(4):899–913
Dagan T, Martin W (2009) Getting a better picture of microbial evolution en route to a network of genomes. Philos Trans R Soc Lond Ser B Biol Sci 364(1527):2187–2196
Darwin C (1860) On the origin of species by means of natural selection, 2nd edn. John Murray, London
Dunne JA, Williams RJ, Martinez ND (2002) Food-web structure and network theory: the role of connectance and size. Proc Natl Acad Sci USA 99(20):12917–12922
Erwin DH (2015) A public goods approach to major evolutionary transitions. Geobiology 13:308–315
Estes JA, Smith NS, Palmisano JF (1978) Sea otter predation and community organization in Western Aleutian Islands, Alaska. Ecology 59(4):822–833
Felsenstein J (2003) Inferring phylogenies. Oxford University Press, Oxford, p 580
Fenollar F et al (2014) Tropheryma whipplei and Whipple’s disease. J Infect 69(2):103–112. https://doi.org/10.1016/j.jinf.2014.05.008
Ge F, Wang LS, Kim J (2005) The cobweb of life revealed by genome-scale estimates of horizontal gene transfer. PLoS Biol 3(10):e316
Gorvel L et al (2010) Tropheryma whipplei, the Whipple’s disease bacillus, induces macrophage apoptosis through the extrinsic pathway. Cell Death Dis 1(4):e34–e34
Goyal A (2018) Metabolic adaptations underlying genome flexibility in prokaryotes. PLoS Genet 14(10):e1007763
Grohmann E, Muth G, Espinosa M (2003) Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev 67(2):277–301
Haggerty LS et al (2014) A pluralistic account of homology: adapting the models to the data. Mol Biol Evol 31(3):501–516
Hahn MW (2018) Molecular population genetics. Oxford University Press, Oxford
Hanage WP (2013) Fuzzy species revisited. BMC Biol 11:41
Hanage WP (2016) Not so simple after all: bacteria, their population genetics, and recombination. Cold Spring Harb Perspect Biol 8(7). https://doi.org/10.1101/cshperspect.a018069
Her HL, Wu YW (2018) A pan-genome-based machine learning approach for predicting antimicrobial resistance activities of the Escherichia coli strains. Bioinformatics 34(13):i89–i95
Hill WG, Robertson A (1966) The effect of linkage on limits to artificial selection. Genet Res 8(3):269–294
Hubner S et al (2019) Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat Plants 5(1):54–62
Jain S, Krishna S (2001) A model for the emergence of cooperation, interdependence, and structure in evolving networks. Proc Natl Acad Sci USA 98(2):543–547
Jones SA et al (2007) Respiration of Escherichia coli in the mouse intestine. Infect Immun 75(10):4891–4899
Karasov TL et al (2018) Arabidopsis thaliana and Pseudomonas pathogens exhibit stable associations over evolutionary timescales. Cell Host Microbe 24(1):168–179.e4
Koonin EV, Wolf YI (2008) Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36(21):6688–6719
Ku C et al (2015) Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524(7566):427–432
Laing C et al (2010) Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics 11:461
Land M et al (2015) Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics 15(2):141–161
Lederberg J, Tatum EL (1946) Gene recombination in Escherichia coli. Nature 158(4016):558
Lukjancenko O, Wassenaar TM, Ussery DW (2010) Comparison of 61 sequenced Escherichia coli genomes. Microb Ecol 60(4):708–720
McInerney JO (2013) More than tree dimensions: inter-lineage evolution’s ecological importance. Trends Ecol Evol 28(11):624–625
McInerney JO, Erwin DH (2017) The role of public goods in planetary evolution. Philos Trans A Math Phys Eng Sci 375(2109). https://doi.org/10.1098/rsta.2016.0359
McInerney JO et al (2011a) The public goods hypothesis for the evolution of life on earth. Biol Direct 6:41
McInerney J, Cummins C, Haggerty L (2011b) Goods-thinking vs. tree-thinking: finding a place for mobile genetic elements. Mob Genet Elem 1(4):1–4
McInerney JO, O’Connell MJ, Pisani D (2014) The hybrid nature of the Eukaryota and a consilient view of life on Earth. Nat Rev Microbiol 12(6):449–455
McInerney JO, McNally A, O’Connell MJ (2017) Why prokaryotes have pangenomes. Nat Microbiol 2:17040
McNally A et al (2016) Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet 12(9):e1006280
Meheust R et al (2018) Formation of chimeric genes with essential functions at the origin of eukaryotes. BMC Biol 16(1):30
Merhej V et al (2011) The rhizome of life: the sympatric Rickettsia felis paradigm demonstrates the random transfer of DNA sequences. Mol Biol Evol 28(11):3213–3223
Nagler M et al (2018) Extracellular DNA in natural environments: features, relevance and applications. Appl Microbiol Biotechnol 102(15):6343
Nanda AM, Thormann K, Frunzke J (2015) Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J Bacteriol 197(3):410–419
Naor A, Gophna U (2013) Cell fusion and hybrids in Archaea: prospects for genome shuffling and accelerated strain development for biotechnology. Bioengineered 4(3):126–129
Nelson-Sathi S et al (2012) Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci USA 109(50):20537–20542
Niehus R et al (2015) Migration and horizontal gene transfer divide microbial genomes into multiple niches. Nat Commun 6:8924
Okasha S (2006) Evolution and the levels of selection. Oxford University Press, Oxford
Paine RT (1969) A note on trophic complexity and community stability. Am Nat 103(929):91–93
Pavlopoulos GA et al (2018) Bipartite graphs in systems biology and medicine: a survey of methods and applications. Gigascience 7(4):1–31. https://doi.org/10.1093/gigascience/giy014
Potera C (1999) Forging a link between biofilms and disease. Science 283(5409):1837–1839
Price MN, Arkin AP (2015) Weakly deleterious mutations and low rates of recombination limit the impact of natural selection on bacterial genomes. MBio 6(6):e01302–e01315
Read BA et al (2013) Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499(7457):209–213
Robins G et al (2007) An introduction to exponential random graph (p∗) models for social networks. Soc Netw 29(2):173–191
Rubino F et al (2017) Divergent functional isoforms drive niche specialisation for nutrient acquisition and use in rumen microbiome. ISME J 11(4):932–944. https://doi.org/10.1038/ismej.2016.172
Sáenz Y et al (2004) Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob Agents Chemother 48(10):3996–4001
Samuelson PA (1954) The pure theory of public expenditure. Rev Econ Stat 36(4):387–389
Savageau MA (1983) Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am Nat 122(6):732–744
Schonknecht G, Weber AP, Lercher MJ (2014) Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. Bioessays 36(1):9–20
Sharp PM et al (1993) Codon usage: mutational bias, translational selection, or both? Biochem Soc Trans 21(4):835–841
Shendure J et al (2017) DNA sequencing at 40: past, present and future. Nature 550(7676):345–353
Sheppard SK, Guttman DS, Fitzgerald JR (2018) Population genomics of bacterial host adaptation. Nat Rev Genet 19(9):549–565
Sherman RM et al (2019) Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat Genet 51(1):30–35
Skiena SS (2008) The algorithm design manual. Springer, London
Steinberger RE, Holden PA (2005) Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl Environ Microbiol 71(9):5404–5410
Stickler DJ (2008) Bacterial biofilms in patients with indwelling urinary catheters. Nat Clin Pract Urol 5(11):598–608
Sunagawa S et al (2015) Structure and function of the global ocean microbiome. Science 348(6237):1261359
Tenaillon O et al (2010) The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8(3):207–217
Tettelin H et al (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci USA 102(39):13950–13955
The Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486(7402):207–214
van Ham RCHJ et al (2003) Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA 100(2):581–586
Vincke E, Boon N, Verstraete W (2001) Analysis of the microbial communities on corroded concrete sewer pipes—a case study. Appl Microbiol Biotechnol 57(5–6):776–785
Wang W et al (2018) Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557(7703):43–49
Welch RA et al (2002) Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA 99(26):17020–17024
Woese CR (2002) On the evolution of cells. Proc Natl Acad Sci 99(13):8742–8747
Yang C et al (2019) Why panmictic bacteria are rare. bioRxiv. https://doi.org/10.1101/385336
Zinder ND, Lederberg J (1952) Genetic exchange in Salmonella. J Bacteriol 64(5):679–699
Acknowledgements
We would like to acknowledge the financial support of the Biotechnology and biological sciences research council (BBSRC) grant number BB/N018044/1. JOM and MOC would like to acknowledge a start-up grant from the School of Life Sciences, University of Nottingham. FJW would like to acknowledge that this project received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 793818.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
McInerney, J.O., Whelan, F.J., Domingo-Sananes, M.R., McNally, A., O’Connell, M.J. (2020). Pangenomes and Selection: The Public Goods Hypothesis. In: Tettelin, H., Medini, D. (eds) The Pangenome. Springer, Cham. https://doi.org/10.1007/978-3-030-38281-0_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-38281-0_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-38280-3
Online ISBN: 978-3-030-38281-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)