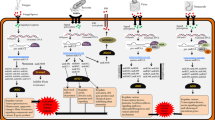
Abstract
MicroRNAs (miRNAs) are small noncoding RNAs involved in post-transcriptional regulation of gene expression in eukaryotes. Given the rapid advancement of sequencing techniques, there has been a dramatic increase in the number of described miRNAs in plants. miRNAs act as master regulators of diverse plant developmental processes and adaptation to environmental stress. Evidence has also emerged on miRNAs involved in antifungal and antibacterial resistance by regulating ETI and PTI responses. Small RNAs, including miRNAs, also mediate cross-kingdom regulation of gene expression in host/pathogen interactions where specific host-derived miRNAs can be transmitted to the pathogen to downregulate genes essential for pathogen virulence. Thus, plants might have adapted cross-kingdom RNA interference mechanisms as part of their battery of immune responses used to arrest pathogen infection. However, although a large number of miRNAs are known to be regulated during pathogen infection, the biological role of the majority of these pathogen-regulated miRNAs has not yet been determined. In this review, we focus on miRNAs for which functional evidence exists on their involvement in the plant response to pathogen infection, mainly fungal pathogens. Elucidation of the function of miRNAs in plant immunity holds great potential for developing novel strategies to improve disease resistance in plants.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Allen E, Xie Z, Gustafson AM et al (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36:1282–1290
Ambros V, Bartel B, Bartel DP et al (2003) A uniform system for microRNA annotation. RNA9:277–279
Arikit S, Zhai J, Meyers BC (2013) Biogenesis and function of rice small RNAs from non-coding RNA precursors. Curr Opin Plant Biol 16:170–179
Axtell MJ (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64:137–159
Axtell MJ, Meyers BC (2018) Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell 30:272–284
Axtell MJ, Westholm JO, Lai EC (2011) Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol 12:221–234
Baldrich P, Kakar K, Siré C et al (2014) Small RNA profiling reveals regulation of Arabidopsis miR168 and heterochromatic siRNA415 in response to fungal elicitors. BMC Genom 15:1083–1099
Baldrich P, Campo S, Wu MT et al (2015) MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol 12:847–863
Baldrich P, Hsing YIC, San Segundo B (2016) Genome-Wide analysis of polycistronic microRNAs in cultivated and wild rice. Genome Biol Evol 8:1104–1114
Barik S, SarkarDas S, Singh A et al (2014) Phylogenetic analysis reveals conservation and diversification of micro RNA166 genes among diverse plant species. Genomics 103:114–121
Baulcombe D (2004) RNA silencing in plants. Nature 431:356–363
Berens ML, Berry HM, Mine A et al (2017) Evolution of hormone signaling networks in plant defense. Annu Rev Phytopathol 55:401–425
Boccara M, Sarazin A, Thiébeauld O et al (2014) The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and Effector-Triggered Immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog 11:e1004814
Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406
Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324(5928):742–744
Bologna NG, Voinnet O (2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol 65:473–503
Borges F, Martienssen RA (2015) The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16:727–741
Boualem A, Laporte P, Jovanovic M et al (2008) MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. 54:876–887
Brant EJ, Budak H (2018) Plant small non-coding RNAs and their roles in biotic stresses. Front Plant Sci 9:1038
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M et al (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190
Budak H, Akpinar BA (2015) Plant miRNAs: biogenesis, organization and origins. Funct Integr Genomics 15:523–531
Cai Q, Qiao L, Wang M et al (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360:1126–1129
Camargo-Ramírez R, Val-Torregrosa B, San Segundo B (2018) MiR858-mediated regulation of flavonoid-specific MYB transcription factor genes controls resistance to pathogen infection in Arabidopsis. Plant Cell Physiol 59:190–204
Campo S, Peris-Peris C, Siré C et al (2013) Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol 199:212–227
Chandran V, Wang H, Gao F et al (2019) miR396-OsGRFs module balances growth and rice blast disease-resistance. Front Plant Sci 9:1999
Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022–2025
Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25:21–44
Chen L, Meng J, He XL et al (2018) Solanum lycopersicum microRNA1916 targets multiple target genes and negatively regulates the immune response in tomato. Plant Cell Environ 42:1393–1407
Chen K, Wang Y, Zhang R et al (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70:1–31
Choi HW, Klessig DF (2016) DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol 16:232–242
Cook DE, Mesarich CH, Thomma BP (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol 53:541–563
Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16:537–552
Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23:431–442
Curaba J, Singh MB, Bhalla PL (2014) miRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot 65:1425–1438
D’Ario M, Griffiths-Jones S, Kim M (2017) Small RNAs: big impact on plant development. Trends Plant Sci 22:1056–1068
De Felippes FF, Schneeberger K, Dezulian T et al (2008) Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA14:2455–2459
Debernardi JM, Rodriguez RE, Mecchia MA et al (2012) Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet 8:e1002419
Demidchik V (2015) Mechanisms of oxidative stress in plants: From classical chemistry to cell biology. Environ Exp Bot 109:212–228
Denancé N, Sánchez-Vallet A, Goffner D, Molina A (2013) Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front Plant Sci 4:155
Ellendorff U, Fradin EF, De Jonge R, Thomma BPHJ (2009) RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J Exp Bot 60:591–602
Fahlgren N, Jogdeo S, Kasschau KD et al (2010) MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22:1074–1089
Gupta OP, Sharma P, Gupta RK, Sharma I (2014) Current status on role of miRNAs during plant–fungus interaction. Physiol Mol Plant Pathol 85:1–7
Hou Y, Zhai Y, Feng L et al (2019) A Phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe 25:153–165
Hua C, Zhao JH, Guo HS (2018) Trans-kingdom RNA silencing in plant–fungal pathogen interactions. Mol Plant 11:235–244
Huang J, Yang M, Lu L, Zhang X (2016) Diverse functions of small RNAs in different plant–pathogen communications. Front Microbiol 7:1552
Islam W, Qasim M, Noman A et al (2018) Plant microRNAs: front line players against invading pathogens. Microb Pathog 118:9–17
Itoh JI, Hibara KI, Sato Y, Nagato Y (2008) Developmental role and auxin responsiveness of Class III Homeodomain Leucine Zipper gene family members in rice. Plant Physiol 147:1960–1975
Jiao J, Peng D (2018) Wheat microRNA1023 suppresses invasion of Fusarium graminearum via targeting and silencing FGSG_03101. J Plant Interact 13:514–521
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Katiyar-Agarwal S, Jin H (2010) Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol 48:225–246
Kim KM, Abdelmohsen K, Mustapic M et al (2017) RNA in extracellular vesicles. Wiley Interdiscip Rev RNA 8:1–10
Knip M, Constantin ME, Thordal-Christensen H (2014) Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet 10:e1004602
Koch A, Biedenkopf D, Furch A et al (2016) An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog 12:e1005901
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) MiRBase: from microRNA sequences to function. Nucleic Acids Res 47:155–162
LaMonte G, Philip N, Reardon J et al (2012) Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 12:187–199
Lee HJ, Park YJ, Kwak KJ et al (2015) MicroRNA844-guided downregulation of Cytidinephosphate Diacylglycerol Synthase3 (CDS3) mRNA affects the response of Arabidopsis thaliana to bacteria and fungi. Mol Plant–Microbe Interact 28:892–900
Lefebvre FA, Lécuyer E (2017) Small luggage for a long journey: transfer of vesicle-enclosed small RNA in interspecies communication. Front Microbiol 8:337
Li Y, Zhang Q, Zhang J et al (2010) Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol 152:2222–2231
Li F, Pignatta D, Bendix C et al (2012) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109:1790–1795
Li Y, Lu YG, Shi Y et al (2014) Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol 164:1077–1092
Li ZY, Xia J, Chen Z et al (2016) Large-scale rewiring of innate immunity circuitry and microRNA regulation during initial rice blast infection. Sci Rep 6:25493
Li Y, Zhao SL, Li JL et al (2017) Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front Plant Sci 8:2
Li Y, Cao XL, Zhu Y et al (2019) Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol 222:1507–1522
Lin S, Chiang SF, Lin W et al (2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147:732–746
Liu H, Wang X, Wang HD et al (2012) Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat Commun 3:1073
Liu J, Cheng X, Liu D et al (2014) The miR9863 family regulates distinct Mlaalleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet 10:e1004755
Liu W, Meng J, Cui J, Luan Y (2017) Characterization and function of microRNA’s in plants. Front Plant Sci 8:2200
Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056
Lu Y, Feng Z, Bian L et al (2010) miR398 regulation in rice of the responses to abiotic and biotic stresses depends on CSD1 and CSD2 expression. Funct Plant Biol 38:44–53
Mallory AC, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14:1035–1046
Mao YB, Cai WJ, Wang JW et al (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25:1307–1313
Meng Y, Shao C (2012) Large-scale identification of mirtrons in arabidopsis and rice. PLoS ONE 7:e31163
Merchan F, Boualem A, Crespi M, Frugier F (2009) Plant polycistronic precursors containing non-homologous microRNAs target transcripts encoding functionally related proteins. Genome Biol 10:R136
Meyers BC, Axtell MJ, Bartel B et al (2008) Criteria for annotation of plant microRNAs. Plant Cell 20:3186–3190
Mi S, Cai T, Hu Y et al (2008) Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133:116–127
Monteiro F, Nishimura MT (2018) Structural, functional, and genomic diversity of plant NLR proteins: an evolved resource for rational engineering of plant immunity. Annu Rev Phytopathol 56:243–267
Navarro L, Dunoyer P, Jay F et al (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436–439
Navarro L, Jay F, Nomura K et al (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science 321:964–967
Nekrasov V, Wang C, Win J et al (2017) Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep 7:482
Nozawa M, Miura S, Nei M (2012) Origins and evolution of microRNA genes in plant species. Genome Biol Evol 4:230–239
Ouyang S, Park G, Atamian HS et al (2014) MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog 10:e1004464
Palatnik JF, Allen E, Wu X et al (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Park YJ, Lee HJ, Kwak KJ et al (2014) MicroRNA400-guided cleavage of Pentatricopeptide Repeat Protein mRNAs renders Arabidopsis thaliana more susceptible to pathogenic bacteria and fungi. Plant Cell Physiol 55:1660–1668
Peláez P, Sanchez F (2013) Small RNAs in plant defense responses during viral and bacterial interactions: similarities and differences. Front Plant Sci 4:343
Peng Y, van Wersch R, Zhang Y (2018) Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol Plant–Microbe Interact 31:403–409
Peris-Peris C, Serra-Cardona A, Sánchez-Sanuy F et al (2017) Two NRAMP6 isoforms function as iron and manganese transporters and contribute to disease resistance in rice. Mol Plant-Microbe Interact 30(5):385–398
Qi T, Guo J, Peng H et al (2019) Host-induced gene silencing: a powerful strategy to control diseases of wheat and barley. Int J Mol Sci 20:206
Qiao Y, Liu L, Xiong Q et al (2013) Oomycete pathogens encode RNA silencing suppressors. Nat Genet 45:330–333
Qin Z, Li C, Mao L, Wu L (2014) Novel insights from non-conserved microRNAs in plants. Front Plant Sci 5:586
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20:3407–3425
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25:2383–2399
Rubio-Somoza I, Weigel D (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16:258–264
Salvador-Guirao R, Baldrich P, Weigel D et al (2018a) The microRNA miR773 is involved in the Arabidopsisimmune response to fungal pathogens. Mol Plant–Microbe Interact 31:249–259
Salvador-Guirao R, Hsing Y, San Segundo B (2018b) The polycistronic miR166k-166h positively regulates rice immunity via post-transcriptional control of EIN2. Front Plant Sci 9:337
Salvador-Guirao R, Baldrich P, Tomiyama S et al (2019) Osdcl1a activation impairs phytoalexin biosynthesis and compromises disease resistance in rice. Ann Bot 123:79–93
Seo JK, Wu J, Lii Y et al (2013) Contribution of small RNA pathway components in plant immunity. Mol Plant–Microbe Interact 26:617–625
Sevilem I, Miyashima S, Helariutta Y (2013) Cell-to-cell communication via plasmodesmata in vascular plants. Cell Adhes Migr 7:27–32
Shahid S, Kim G, Johnson NR et al (2018) MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553:82–85
Shivaprasad PV, Chen HM, Patel K et al (2012) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24:859–874
Skopelitis DS, Hill K, Klesen S et al (2018) Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat Commun 9:3107
Song Y, Thomma BPHJ (2018) Host-induced gene silencing compromises Verticillium wilt in tomato and Arabidopsis. Mol Plant Pathol 19:77–89
Song X, Li Y, Cao X, Qi Y (2019) MicroRNAs and their regulatory roles in plant–environment interactions. Annu Rev Plant Biol 70:489–525
Soto-Suarez M, Baldrich P, Weigel D et al (2017) The Arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens. Sci Rep 7:44898
Staiger D, Korneli C, Lummer M, Navarro L (2013) Emerging role for RNA-based regulation in plant immunity. New Phytol 197:394–404
Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18:2051–2065
Torres MA, Jones JDG, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141:373–378
Tsikou D, Yan Z, Holt DB et al (2018) Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362:233–236
Tsuda K, Somssich IE (2015) Transcriptional networks in plant immunity. New Phytol 206:932–947
Van Peer G, Lefever S, Anckaert J et al (2014) miRBase tracker: keeping track of microRNA annotation changes. Database (Oxford), vol 2014
Vatén A, Dettmer J, Wu S et al (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21:1144–1155
Vaucheret H (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20:759–771
Vazquez F, Blevins T, Ailhas J et al (2008) Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res 36:6429–6438
Wang Y, Cheng X, Shan Q et al (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32:947–951
Wang M, Weiberg A, Lin FM et al (2016a) Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat Plants 2:16151
Wang F, Wang C, Liu P et al (2016b) Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 11:e0154027
Wang B, Sun Y, Song N et al (2017a) Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol 215:338–350
Wang M, Thomas N, Jin H (2017b) Cross-kingdom RNA trafficking and environmental RNAi for powerful innovative pre- and post-harvest plant protection. Curr Opin Plant Biol 38:133–141
Wang Z, Xia Y, Lin S et al (2018) Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J 95:584–597
Wang J, Mei J, Ren G (2019) Plant microRNAs: biogenesis, homeostasis, and degradation. Front Plant Sci 10:360
Weiberg A, Wang M, Lin FM et al (2013) Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342:118–123
Weiberg A, Wang M, Bellinger M, Jin H (2014) Small RNAs: a new paradigm in plant–microbe interactions. Annu Rev Phytopathol 52:495–516
Wilson RA, Talbot NJ (2009) Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol 7:185–195
Yu Y, Jia T, Chen X (2017) The ‘how’ and ‘where’ of plant microRNAs. New Phytol 216:1002–1017
Zhai J, Jeong DH, de Paoli E et al (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25:2540–2553
Zhang X, Zhao H, Gao S et al (2011) Arabidopsis Argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell 42:356–366
Zhang D, Liu M, Tang M et al (2015) Repression of microRNA biogenesis by silencing of OsDCL1 activates the basal resistance to Magnaporthe oryzae in rice. Plant Sci 237:24–32
Zhang C, Ding Z, Wu K et al (2016a) Suppression of jasmonic acid-mediated defense by viral-inducible microRNA319 facilitates virus infection in rice. Mol Plant 9:1302–1314
Zhang T, Zhao YL, Zhao JH et al (2016b) Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat Plants. 2:16153
Zhang X, Bao Y, Shan D et al (2018) Magnaporthe oryzaeinduces the expression of a microRNA to suppress the immune response in rice. Plant Physiol 177:352–368
Zhu QH, Spriggs A, Matthew L et al (2008) A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res 18:1456–1465
Zipfel C (2013) Combined roles of ethylene and endogenous peptides in regulating plant immunity and growth. Proc Natl Acad Sci USA 110:5748–5749
Acknowledgements
This work was supported by the Ministerio de Ciencia, Innovación y Universidades–Agencia Estatal de Investigación/FEDER (BIO2015-67212-R, RTI2018-101275-B-100, and BIO2017-92113-EXP). We also acknowledge support from the CERCA Programme (‘Generalitat de Catalunya’), and MINECO (‘Severo Ochoa Programme for Centres of Excellence in R&D’ 2016-2019, SEV-2015-0533).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Bundó, M., Campo, S., San Segundo, B. (2020). Role of microRNAs in Plant–Fungus Interactions. In: Miguel, C., Dalmay, T., Chaves, I. (eds) Plant microRNAs. Concepts and Strategies in Plant Sciences. Springer, Cham. https://doi.org/10.1007/978-3-030-35772-6_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-35772-6_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-35771-9
Online ISBN: 978-3-030-35772-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)