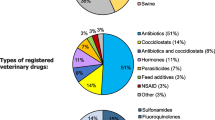
Abstract
Despite a large and rapidly growing volume of research activity and output, primarily on the biological bases of antimicrobial resistance (AMR), epidemiological understanding of the causal mechanisms at play behind the apparent recent global rise in prevalence of AMR has, arguably, progressed very little. Despite this inconvenient fact, political imperative and expedience, among other drivers, have given substantial impetus to an interventionist approach against what are considered to be the culprits for the apparent growing prevalence of AMR and its impacts. Concern about the rise in prevalence of microbial infections that are resistant to therapeutic agents designed to kill them has arisen almost exclusively in relation to human health. (Public awareness and concern about antihelmintic resistance, for which the impacts are much more substantial for animal health, at least in developed temperate countries, are trivial by comparison). Nevertheless, antimicrobial drugs have been, and are, widely used in animal health and production throughout the world, and the contribution of this diverse usage to the ‘global AMR problem’ has historically been controversial. There is growing acceptance, notwithstanding the limitations in causal understanding noted previously, of AMR as an ecological problem of competing populations of microorganisms experiencing both natural and anthropogenic selection pressures in compartments that transcend species and other boundaries. Typifying what is described as a ‘One Health’ problem, AMR is therefore considered to be most amenable to conjoint mitigation efforts in all compartments: i.e. interventions in human health, animal health, food and the environment in a coherent manner.
In animals, this calls into question the motivations and practices for antimicrobial drug usage, the majority of which are justified on the basis of promoting animal health and welfare and securing a food supply for a growing human population. Not surprisingly, there are great differences in animal husbandry and food demand, and in availability, access and regulation of antimicrobial usage in animals, and in surveillance of AMR, which are likely to be starkest between developed and developing countries. Thus, it is unlikely that the impacts of AMR, and the impacts of efforts to mitigate AMR that are directed to the ‘animal compartment’ of the ecosystem, will be felt equally across the world.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction: Evolutionary History of Antimicrobial Resistance as a Natural Phenomenon
Antimicrobial resistance has occurred as a natural phenomenon for millennia, as a response to inhibitory substances produced by microbial populations competing for resources in different ecosystems (Hall and Barlow 2004). Human discovery of the existence of these substances was exploited in the early twentieth century leading to the development of the first antibacterial drugs for therapeutic use. The rapid discovery and development of both natural and synthetic antimicrobial drugs (active against bacteria, fungi, viruses, and protozoa), and their widespread use to treat and prevent human and animalinfectious diseases, took place throughout the latter half of the twentieth century. As predicted by Alexander Fleming himself, as therapeutic use of antibiotics grew there closely followed the emergence of untreatable strains of organisms that had hitherto responded to treatment.
Any microorganism that isn’t inhibited or killed by appropriate, effective antimicrobials is classified as resistant (Ridge et al. 2011). This phenomenon is now widely explained in terms of a ‘selection pressure’ being exerted on the populations of microorganisms which are exposed to antimicrobial agents. Such microbial populations are usually comprised of an almost unimaginable number and diversity of individual organisms, amongst which the target ‘pathogen’ population for antimicrobial therapy may constitute only a fraction of those exposed to the agent. Under these circumstances, those organisms susceptible to the agent are inactivated, and cease to compete for resources, and those that are equipped with mechanism(s) to resist the effects of the agent thrive through access to the resources no longer consumed by the inactivated organisms (Levin et al. 2000). In the case of therapeutic use of antimicrobial agents, if the organisms equipped with mechanism(s) to resist the effects of the agent are members of the ‘pathogen’ population, the result is likely to be treatment failure and prolonged clinical disease for the patient. If the organisms equipped with mechanism(s) to resist the effects of the agent are members of the non-pathogen population (usually referred to as commensals), the effect is to select for populations that carry resistance mechanisms, but is unlikely to result in treatment failure at that time. Nevertheless, and especially for bacteria, many of the mechanisms to resist the effects of antimicrobial agents are coded for by genes carried on transmissible genetic elements which can be passed between organisms of the same or different species or genera. Thus, selection for resistance in members of the non-pathogen population could subsequently lead to transfer to ‘pathogen’ populations and increased likelihood of subsequent treatment failure.
2 Drug Resistance as an Animal or Public Health Concern
Treatment failure and prolonged and/or more serious clinical disease for the patient are probably much more widely recognized consequences of drug resistance in human than in veterinary medicine. In efforts to mitigate these principally public health effects of antimicrobial resistance in the developed world, much attention has been focussed on human healthcare settings, particularly hospitals and care homes (Edwards et al. 2012). Efforts to improve infection prevention and control (IPC) in these settings are paramount, because they are needed not only to reduce the transmission between people of infectious agents likely to require drug therapy, but also the transmission of antimicrobial resistance genes. Principles of IPC form the mainstays of current strategies to combat antimicrobial resistance and are coupled with measures to promote ‘better’ prescribing of antimicrobial agents through guidelines and audit, and initiatives to educate the healthcare profession and the public. Judicial antimicrobial use (i.e. “better prescribing”) is based on evidence which supports using a particular agent against a particular organism in a particular patient for a particular reason and period of time (e.g. British Veterinary Association 2015).
The contribution and nature of veterinary use of antimicrobial agents to the problem of antimicrobial resistance on a global scale has been a controversial issue for many years, with conflicting, polarized views espoused by different respected research groups (Aarestrup et al. 2000; Collignon 2013; Mather et al. 2013; van Bunnik and Woolhouse 2017). Much has been made of the widespread and varied use of antimicrobial agents in veterinary medicine and the potential for this to contribute to the (largely) public health problem posed by antimicrobial resistance. In this, it is noteworthy that, with few exceptions, relatively little is made of treatment failure and prolonged and/or more serious clinical disease for animal patients. The use of the broad term ‘antimicrobial’ has been unhelpful in this regard as it is inclusive of agents such as anti-coccidials, which are important for animal health and food security, but have no effect on other organisms (such as bacteria or fungi) which drive resistance in humans (Mendelson et al. 2017). Mendelson et al. (2017) argue for more precision in the language around drug-resistant infections, and for more specific terms such as “antibiotic” or “antifungal” to be used in preference to ‘“antimicrobials” when referring to medicines against a specific type of organism.
There is a growing acceptance that drug-resistant infections are a ‘One Health’ problem that transcends species (and other) barriers (Karesh et al. 2012). Expanding on this, it is obvious that agents that carry resistance determinants exist very well and evolve outside animal or human hosts. There is increasing recognition of the need to consider environmental reservoirs, and inanimate vehicles of transmission, not least among which are foods of animal origin – home produced and imported – and integrate these into thinking about the ecology and epidemiology of resistance (Wellington et al. 2013). Resistance genes may spread directly from people to animals and from animals to people through food-borne and environmental contamination (via wastewater, soil and the spread of contaminated manure from livestock and wild animals) (Casey et al. 2013; Kim et al. 2013; Davis and Rutkow 2012; Johnson and Becker 2010; Cantas et al. 2013; Roe and Pillai 2003; Soonthornchaikul and Garelick 2009; Levy et al. 1976; Thanner et al. 2016; Literak et al. 2011). However, the relative importance of these routes of transmission is uncertain; exposure is complex and not unequivocally in one direction (Carlet et al. 2011; Mather et al. 2013; Zhu et al. 2013). Additionally, there are other epidemiological pathways (such as transmission of resistance via irrigation and waste-water, plant production and the disposal or presence of disinfectants and heavy metals in the environment) that have been shown to be associated with the emergence of drug resistance. These are not well researched due to the lack of analytical methods to monitor contaminants in waste, surface and drinking water and soil (Thanner et al. 2016). Thus, the epidemiological drivers associated with the selection for and against resistant organisms in animals are unlikely to be different to the causal mechanisms believed to exist in humans or the environment. There are important caveats to this assertion: (1) many parts of the relevant ecosystem are ignored by surveillance approaches adopted to date, notably the environment and food (home produced and imported), (2) it is difficult to partition antibiotic resistance into that which arises naturally from bacterial competition in various ecological niches and that which is selected for anthropogenically through therapeutic or other use of drugs and/or biocides. Of course, the relative intensity of the various selection pressures for the emergence of drug-resistant infections is itself driven in part by broader socio-economic issues (e.g. poverty, sanitation, hygiene and public health resources) which are harder to quantify and even more difficult for governments to address. These caveats arise due to the limitations of the evidence provided by surveillance for drug-resistant infections, which has largely focused on trying to compare observations of antibiotic resistance in bacterial isolates from human patients to those of antibiotic resistance in animal populations (often using different methods of antimicrobial resistance determination) in developed countries.
High rates of drug-resistant infections are found in densely populated, developing countries where there is corruption and unreliable enforcement of laws and regulations pertaining to the practice of human and veterinary medicine (Collignon et al. 2015). Individuals who are exposed to resistant bacteria or fungi in these hotspots through international travel and medical tourism can subsequently import resistance into other countries, resulting in rapid global spread (World Health Assembly Resolution 51.9 1998).
There is still a need to convince some sectors that this is a shared problem with shared accountability and shared responsibility, but that principle is pretty much implicit in nationally and internationally agreed accords (e.g. Department of Health 2013; O’Neill 2016). Based on experience from the UK, plans to implement the recommendations of such accords, whilst all claiming to be taking a ‘One Health’ approach, can vary even among devolved administrations. In this sense, creation of a bespoke ‘One Health’ agency, with appropriately balanced multidisciplinary representation (and buy-in and trust), acting in a collegiate manner to coordinate a collective and coherent response seems intuitively more likely to succeed than simply hoping that separate agencies will be able to work in parallel towards a common aim without ‘funding the arrows’ (Campbell 2006).
In conclusion, considering the epidemiology of drug-resistance in animals in isolation is missing the point.
3 Antimicrobial Use in Animals
Similar to antimicrobial use in humans, there is substantial variation throughout the world in the availability, regulation, control and administration of antimicrobials to animals. Whilst these factors themselves are not expected to alter the postulated causal mechanisms by which drug use affects drug resistance in microorganisms, they are likely to modify the extent to which these mechanisms have the opportunity to act. Simply put, it appears that greater usage of particular antimicrobial drugs, in a relevant time and place, is positively correlated with greater proportions of isolates tested for susceptibility in that time and place being designated as resistant. Thus, greater control over the quality and supply of antimicrobial drugs and greater regulation and professionalism over their administration should correlate with reduced proportions of resistant isolates.
It is worthy of note that this general observation appears also to be true of anthelmintic drugs, although the ecology of macroparasites is of course different to microbes. However, in the developed world, the problem of anthelmintic resistance is more of an immediate issue for animals than humans, which means that it has received much less attention from a public health point of view. Nevertheless, sophisticated guidance on control of parasitic infestations, based on understanding their ecology, and seeking to preserve the efficacy of anthelmintic drugs, are seen as critical to future food security (e.g. see Sustainable Control of Parasites in Sheep (http://www.scops.org.uk/)).
A range of antimicrobial drugs is used widely to treat disease in domestic animals through a large and internationally varying number of preparations and through a number of different routes of administration. In most developed countries of the world, the classes of antimicrobial drug available for animal administration, and the preparations and routes by which they can be administered, have to be specifically licensed for animal use and are highly regulated, especially for animal species likely to enter the human food chain. In these countries, there are also strictly enforced ‘withdrawal periods’ which define for how long an animal must be ‘off’ treatment before products form that animal can be used for human consumption. In some parts of the world, so-called ‘off-label’ use (i.e. an unlicensed product and/or an unlicensed route of administration) is sometimes permitted under derogation for the treatment of companion animals. Lists of critically important antimicrobials (CIAs) in humans and animals have been agreed, and are periodically updated, by the OIE and WHO and others, and the use of these drugs should be restricted to treating infections that have been demonstrated to be susceptible (and resistant to less important, so-called first-line, drugs) (OIE list of antimicrobials of veterinary importance 2007; WHO list of Critically Important Antimicrobials (CIA) 2017). In developing countries, due to limitations in infrastructure, such regulations, where they exist, are much more difficult to make effective. Coupled with a very high demand due to a high disease burden and often high population densities, and issues such as uncontrolled, unauthorized markets and counterfeit drugs, the conditions appear likely to favour intense selection for antimicrobial resistance and subsequent dissemination, though there are few reliable data to provide firm evidence of this.
Much of the controversy around use of antimicrobials in animals has concentrated on their use as growth promoters. It has been considered that routine addition of some antimicrobial compounds (usually antibacterial drugs and often at sub-therapeutic concentrations) to livestock feed increases food conversion efficiency by more than enough to outweigh the cost of adding the drugs, though this is disputed (Graham et al. 2007). However, other agents, such as anti-coccidials used by the poultry sector, are also included in this broad classification (Mendelson et al. 2017). Use of antibiotic growth promoters is considered by many to be particularly undesirable due to the selection pressure being applied in an almost unrelenting way to the populations of microorganisms colonizing these animals, especially in instances in which drugs are used at sub-therapeutic concentrations, as this is thought to select more strongly for antimicrobial resistance. Much research has sought to investigate the impact of the use of antimicrobials for growth promotion and has been interpreted by most as demonstrating a positive association with increased prevalence of resistant organisms in exposed microbial populations (O’Neill (2015)). On the basis of this evidence, their use as growth promoters has been banned in many parts of the world, notably Europe, yet still persists in others.
In the EU, “the use of agents from classes which are or may be used in human or veterinary medicine (i.e. where there is a risk of selecting for cross-resistance to drugs used to treat bacterial infections)” as growth promoters has not been permitted since 2006 (Regulation (EC) No 1831/2003/EC) and withdrawal periods for antimicrobial use prior to animal slaughter are designed to ensure that there are no antimicrobial residues in food. In the USA, non-therapeutic use is still widespread in industrial farming. The USA FDA has historically been slow to respond to calls to reduce antimicrobial use and unwilling to exert its authority over the antimicrobial approval process (see Natural Resources Defense Council, et al. v. United States Food and Drug Administration, et al.). Until recently, the FDA implemented a voluntary approach to antimicrobial conservation that encouraged drug companies to withdraw approvals for antimicrobials for non-therapeutic use and replace them with approvals for other uses such as chemoprophylaxis (USDA 2012a, b). However, this policy has had little real effect on antimicrobial usage because, in many cases, the doses and durations of drug use for chemoprophylaxis and growth promotion are the same (Outterson 2014).
In emerging economies, such as the BRICS and MINT countries, it is anticipated that non-therapeutic use of antibiotics in animals may exponentially increase because of increasing intensification of agriculture/aquaculture, high prevalences of production and endemic diseases (which are likely to be better controlled in other countries) and lack of resources to ensure appropriate governance over antimicrobial use (Carlet et al. 2011; Van Boeckel 2015). In some of these countries, AMR and antimicrobial conservation aren’t on the “political agenda” at all (Grace 2015 at p 11–12) because addressing other issues such as poverty, starvation, malnutrition through (un)sustainable livestock and farmed fish production are more urgent for the current population. The impact of antimicrobial use on accessibility of animal food sources hasn’t been quantified for most countries, partly due to the variability and uncertainty regarding the quantities of antimicrobials used even in similarly intensive systems elsewhere (Rushton et al. 2014), and there is no agreement on the desired levels of antimicrobial consumption. Even if an enforced ban on the non-therapeutic use of antimicrobials were to be introduced, in some places, the absence of national R&D investment means that there are few alternative mechanisms (such as vaccines) to concurrently improve animal husbandry and avoid production losses which could paradoxically increase AMR prevalence through off-label or unprescribed use of poor quality or counterfeit antimicrobials. The relative costs of not using antimicrobials on the security of the global food supply and the success of the Sustainable Development Goals (in eradicating poverty and hunger in the current generation) have not been compared to AMR treatment failure in people (in future generations). Although developed countries arguably have sufficient means to assist developing countries address some of these issues, they have so far focussed on their own national priorities in order to achieve wider international societal benefits (Clift 2013).
Distinction is made in veterinary medicine (at least in the developed world) between prophylactic (administering antimicrobial drugs to prevent anticipated infection) and metaphylactic use of antimicrobials (administering antimicrobial drugs to clinically well members of a population in contact with an index case of infectious disease to prevent anticipated infection), which is not recognized in human medicine. Prophylaxis in veterinary medicine is criticized by many as an excuse for poor infection prevention and control (often referred to as ‘biosecurity’ in animal production) in a particular animal husbandry system, but presents ethical dilemmas to veterinarians who struggle with the notion of withholding treatment in the face of what is considered to be almost inevitable disease and associated welfare compromise. In many instances, the clinical use of antimicrobials in animals is empirical and sensitivity testing of an isolate of the putative causative agent of infection is not carried out. The reasons for this are largely to do with cost and expediency in starting treatment to improve the clinical condition of the patient. Thus, data that characterize the resistance profiles of clinical isolates from animals represent a very small proportion of the putative infections treated by veterinarians, and their use for epidemiological purposes often appears to overlook this fact. However, this is also true of human medicine.
The notion that antimicrobial resistant organisms somehow ‘arise’ in animals, driven by selection pressures applied by veterinary use of antimicrobials, and pass to humans, principally through the food chain, has been a popular model of antimicrobial resistance ‘acquisition’ for some time. However, some recent research shows that transmission is likely to occur in both directions between animals and people directly and indirectly through the environment and various fomites (Mather et al. 2013). As discussed in the introduction, it is probably more reasonable to think of animals and people as inextricably linked ‘samplers’ of a shared environmental pool of organisms subject to different selection pressures in different compartments.
4 Surveillance for Antimicrobial Resistance in Animals
‘Better’ surveillance for antimicrobial resistance in animals is a more or less universal feature of international concordats, calls to action and other declarations on antimicrobial resistance (Department of Health 2013; O’Neill 2016). As the bedrock of epidemiology, surveillance activity, and the intelligence generated by it, offers the greatest potential to understand fully the causal relationships at play within the complex landscape ecology underlying the antimicrobial resistance ‘phenomenon’ (Singer et al. 2006). Nevertheless, there are a great many limitations, often overlooked, that can apply to data derived from surveillance of infections in animals (and people) for the purposes of exploring causal relationships of relevance to the emergence and spread of antimicrobial resistance. Chief among these is the question of how well the isolate(s) derived from the sample(s) collected from the individual(s) and/or their environment(s) under study represent the actual nature and dynamics of the interactions between host and microbial populations and the selection pressures experienced by them. Confounding this, in many instances, is a lack of standardization of the microbiological methods applied and inconsistent definitions of how antimicrobial sensitivity or lack of it is defined for individual drug/bug combinations derived from different species (animals and humans).
The World Health Organization (WHO) has led international efforts to develop action plans to monitor and reduce drug-resistant infections (WHO Strategy 2001). However, without a legally-binding mandate, it has been difficult implement this strategy within Member States. Indeed, between 1998 and 2015 (when the WHO first published its global action plan), there have been at least seven World Health Assembly (WHA) Resolutions promoting surveillance of drug-resistant infections, but still no internationally coordinated or standardized AMRsurveillance strategy within either the human or veterinary sectors. Although collaboration between the WHO, Food and Agricultural Organization of the United Nations (FAO) and World Organization of Animal Health (OIE) is improving coordination between human and animalsurveillance, within individual Member States the two sectors remain distinct and regulated independently of one another. Surveillance, if undertaken at all, has hitherto been implemented separately by each sector, without harmonization or standardization of approach.
Existing national action plans within Member States describe broad strategic aims to mitigate, and reduce current rates of drug-resistant infections and resultant treatment failures. These include: improvements in infection prevention and control, education and training initiatives, optimization of prescribing practices (particularly in animals with respect to critically important antimicrobials) and incentives to innovate new, effective therapies. For the most part, these aims have not been translated yet into specific actions with explicit timescales for delivery or agreed outcome metrics. In some countries, technical and financial constraints, such as lack of existing public health infrastructure, access to diagnostic technologies and changing public attitudes towards public health mean that surveillance is poor (STAG-AMR 2013; WHO 2011). Accurate inferences (and/or between-country comparisons) about antibiotic or antifungal consumption rates aren’t always possible because of the scarcity and variability of available information on consumption, species treated, routes of administration, dose rate, and pharmaceutical costs (Rushton et al. 2014 at p 11). Other important differences, such as husbandry practices, diversity of available drugs and prescribing habits of veterinarians, mean that comparisons of antimicrobial use based on animal demographics are potentially misleading. For example, Chile is the second largest producer of farmed salmon and the only important developing country producer. However, it uses around 300 times more antibiotics than the largest salmon producer, Norway because Norway has the resources to access and implement vaccination and alternative husbandry measures instead of antimicrobials to control diseases (Grace 2015 at page 11). Thus, it is currently not possible to accurately chart progress towards AMR reduction or identify early-warning performance indicators for actions that aren’t effective in achieving these aims.
Despite these limitations, surveillance and research data generated, principally in the developed world, are subject to ever more sophisticated epidemiological analyses to infer risk factors and causal relationships for resistance emergence, persistence and spread. Entrenched in this aim is the notion that population patterns of drug-resistant infections can be explained by a complex web of multiple interconnected factors, which if identified, can be used to inform and target interventions to improve public health (Krieger 1994). Increasingly, these analyses seek to be comparative and inferential about the impacts of antimicrobial use in one species (animals or man) on antimicrobial resistance in the other, as the ‘One Health’ construct of drug resistance becomes more widely accepted. However, caution is required in the interpretation of these findings. ‘Causal webs’ are not unbiased; they are necessarily hierarchical, focusing “attention on risk factors closest to the outcome under inspection”, inevitably prioritizing biological factors (amenable to medical or veterinary intervention) over other broader social or environmental determinants which could be addressed through social action (Krieger 1994; Thanner et al. 2016). For example, addressing a lack of access to education, sanitation and adequate healthcare (including infection prevention and control, scarcity of new antibiotics, poor prescribing practices and absence of concurrent diagnostic tools to ensure appropriate treatment) will affect rates of disease, which will influence subsequent amounts of antimicrobial used (Buckland Merrett 2013). Reduction of antimicrobial use may reduce selection pressure for resistance, but at the same reduces the numbers of antimicrobial ‘customers’ and thus incentives to innovate new drugs. Equally, economic strategies that are driven by sales rewards will inevitably conflict with principles of conservation and IPC. Understanding of the epidemiology of AMR (and this broader socio-economic and ecological ‘web of causation’) and an appreciation for the gaps in communication and coordination between stakeholders and regulators involved across multiple sectors and disciplines at each of these foci is therefore necessary to ensure policy decisions are robust and ethical.
5 Summary and Conclusion
The epidemiology of antimicrobial resistance in animals seems very unlikely to be extricable or distinct from that in people. A powerful epidemiological model of the causation and dissemination of antimicrobial resistance as a feature of microbiological ecology among a very complex web of interconnected host and environmental compartments has emerged and gained widespread acceptance under the construct of ‘One Health’. However, the limitations in the quality of the data that have been used to build this model leave room for uncertainty about its validity. In addition, there needs to be ‘thinking time and space’ to consider and account for cognitive biases in such models and to incorporate socio-economic modifiers of such biological models in order to inform efficient and effective measured interventions. As rapidly developing scientific advances offer the potential to improve the quality (e.g. WGS) and representativeness (e.g. through properly and purposively designed comparative surveillance programmes) of data, it is to be hoped that epidemiological inference will be able to keep pace and offer better, and what yet may be surprising, insights into the problem of antimicrobial resistance and ways in which it might be tackled.
References
Aarestrup, F.M., Y. Agerso, P. Gerner-Schmidt, M. Madsen, and L.B. Jense. 2000. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers and pigs in Denmark. Diagnostic Microbiology and Infectious Disease 27: 127–137.
British Veterinary Association. 2015. Responsible use of antimicrobials in veterinary practice. Available from: https://www.bva.co.uk/uploadedFiles/Content/News,_campaigns_and_policies/Policies/Medicines/responsible-use-of-antimicrobials-in-veterinary-practice.pdf.
Buckland Merrett, G.L. 2013. Tackling antibiotic resistance for greater global health security. Centre on Global Health Security. Available from: http://www.chathamhouse.org/sites/files/chathamhouse/public/Research/Global%20Health/1013bp_antibioticresistance.pdf.
Campbell, A. 2006. The Australian natural resource management knowledge system. Available from: http://lwa.gov.au/products/pr061081.
Cantas, L., S.Q.A. Shah, L.M. Cavaco, C.M. Manaia, F. Walsh, M. Popowska, H. Garelick, H. Bürgmann, and H. Sørum. 2013. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Frontiers in Microbiology 4: 96. https://doi.org/10.3389/fmicb.2013.00096.
Carlet, J., P. Collignon, D. Goldmann, H. Goossens, Gyssens ICJ, S. Harbarth, V. Jarlier, S.B. Levy, B. N’Doye, D. Pittet, R. Richtmann, W.H. Seto, J.W.M. van der Meer, and A. Voss. 2011. Society’s failure to protect A precious resource: Antibiotics. The Lancet 378 (9788): 369–371.
Casey, J.A., F.C. Curriero, F.C. Cosgrove, K.E. Nachman, and B.S. Schwartz. 2013. High-density livestock operations, crop field application of manure, and risk of community-associated methicillin-resistant Staphylococcus Aureus infection. Journal of the American Medical Association of Internal Medicine 173 (21): 1980–1990. https://doi.org/10.1001/jamainternmed.2013.10408.
Clift, C. 2013. The UK’s new antimicrobial resistance strategy. Available at https://www.chathamhouse.org/media/comment/clift/amr (last accessed 31 July 2015).
Collignon, P. 2013. Superbugs in food: A severe public health concern. The Lancet Infectious Disease 13 (8): 641–643.
Collignon, P., A. Prema-chandra, S. Senanayake, and F. Khan. 2015. Antimicrobial resistance: The major contribution of poor governance and corruption to this growing problem. PLoS One 10 (3): e0116746. https://doi.org/10.1371/journal.pone.0116746.
Davis, M.F., and L. Rutkow. 2012. Regulatory strategies to combat antimicrobial resistance of animal origin: Recommendations for a science-based US approach. Tulane Environmental Law Journal 25: 327–387.
Department of Health. 2013. UK five year antimicrobial resistance strategy 2013 to 2018. Available from: https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/244058/20130902_UK_5_year_AMR_strategy.pdf.
Edwards, R., E. Charani, N. Sevdalis, B. Alexandrou, E. Sibley, D. Mullett, H.P. Loveday, L.N. Drumright, and A. Holmes. 2012. Optimisation of infection prevention and control in acute health care by use of behaviour change: A systematic review. Lancet Infectious Diseases 12: 318–329. https://doi.org/10.1016/S1473-3099(11)70283-3.
Grace, D. 2015. Review of evidence on antimicrobial resistance and animal agriculture in developing countries. ILRI Available from: http://www.evidenceondemand.info/review-of-evidence-on-antimicrobial-resistance-and-animal-agriculture-in-developing-countries.
Graham, J.P., J.J. Boland, and E. Silbergeld. 2007. Growth promoting antibiotics in food animal production: An economic analysis. Public Health Reports 122: 79–87.
Hall, B.G., and M. Barlow. 2004. Evolution of the serine beta-lactamases: Past, present and future. Drug Resistance Updates 7: 111–123. https://doi.org/10.1016/j.drup.2004.02.003.
Johnson, R., and G. Becker. 2010. US-Russia meat and poultry trade issues. Washington, DC: Congressional Research Service for Congress. Available from: http://nationalaglawcenter.org/wp-content/uploads/assets/crs/RS22948.pdf.
Karesh, W.B., A. Dobson, J.O. Lloyd-Smith, J. Lubroth, M.A. Dixon, M. Bennett, S. Aldrich, T. Harrington, P. Formenty, E.H. Loh, C.C. Machalaba, M.J. Thomas, and D.L. Heymann. 2012. Ecology of zoonoses: Natural and unnatural histories. The Lancet 380: 1936–1945.
Kim, B.F., L.I. Laestadius, R.S. Lawrence, R.P. Martin, S.E. Mckenzie, K.E. Nachman, T.J.S. Smith, and P. Truant. (2013). Industrial food animal production in America: Examining the impact of the Pew Commission’s Priority Recommendations. John’s Hopkins Centre for a Liveable Future. Available at http://www.jhsph.edu/research/centers-and-institutes/johns-hopkins-center-for-a-livable-future/_pdf/research/clf_reports/CLF-PEW-for%20Web.pdf.
Krieger, N. 1994. Epidemiology and the web of causation: Has anyone seen the spider? Social Science Medicine 39 (7): 887–903.
Levin, B.R., V. Perrot, and N. Walker. 2000. Compensatory mutations, antibiotic resistance and population genetics of adaptive evolution in bacteria. Genetics 154: 985–997.
Levy, S.B., G.B. FitzGerald, and A.B. Macone. 1976. Changes in intestinal flora of farm personnel after introduction of a tetracycline-supplemented feed on a farm. New England Journal of Medicine 295: 583–588. https://doi.org/10.1056/NEJM197609092951103.
Literak, I., R. Petro, M. Dolejska, E. Gruberova, H. Dobiasova, J. Petr, et al. 2011. Antimicrobial resistance in fecal Escherichia coli isolates from healthy urban children of two age groups in relation to their antibiotic therapy. Antimicrobial Agents and Chemotherapy 55: 3005–3007. https://doi.org/10.1128/AAC.01724-10.
Mather, A.E., S.W.J. Reid, D.J. Maskell, J. Parkhill, M.C. Fookes, S.R. Harris, D.J. Brown, J.E. Coia, M.R. Mulvey, M.W. Gilmour, L. Petrovska, E. de Pinna, M. Kuroda, M. Akiba, H. Izumiya, T.R. Connor, M.A. Suchard, P. Lemey, D.J. Mellor, D.T. Haydon, and N.R. Thomson. 2013. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 341: 1514–1517. https://doi.org/10.1126/science.1240578.
Mendelson, M., M. Balasegaram, T. Jinks, C. Pulcini, and T. Sharland. 2017. Antibiotic resistance has a language problem. Nature 545 (7652): 23–25.
Natural Resources Defense Council. et al. 2012a. v. United States Food and Drug Administration, et al. 884 F.Supp.2d 127 (S.D.N.Y.).
Natural Resources Defense Council. et al. 2012b. v. United States Food and Drug Administration, 872 F.Supp.2d 318 (S.D.N.Y.).
O’Neill, J. 2015. Antimicrobials in agriculture and the environment: Reducing unnecessary use and waste. Available from: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf.
———. 2016. Tackling drug-resistant infections globally: Final report and recommendations. Available from: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf.
OIE list of antimicrobials of veterinary importance. 2007. Available from: https://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/AMR/A_OIE_List_antimicrobials_July2019.pdf
Outterson, K. 2014. New business models for sustainable antibiotics. Centre on global health security working group papers 1–31. Available from: http://www.chathamhouse.org/sites/files/chathamhouse/public/Research/Global%20Health/0214SustainableAntibiotics.pdf.
Regulation (EC) No 1831/2003/EC Of The European Parliament and of the Council of 22 September 2003 On additives for use in animal nutrition, replacing directive 70/524/EEC on additives in feeding-stuffs. 2003. Official Journal of the European Union L 268: 0029–0043.
Ridge, K.W., K. Hand, M. Sharland, I. Abubakar, and D.M. Livermore. 2011. Antimicrobial resistance. In The annual report of the chief medical officer volume 2, Infections and the rise of antimicrobial resistance. Available at https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/138331/CMO_Annual_Report_Volume_2_2011.pdf
Roe, M.T., and S.D. Pillai. 2003. Monitoring and identifying antibiotic resistance mechanisms in bacteria. Poultry Science 82: 622–626.
Rushton, J., J. Pinto Ferreira, and K.D. Stärk. 2014. Antimicrobial resistance: The use of antimicrobials in the livestock sector, OECD Food, Agriculture and Fisheries Papers No. 68. Paris: OECD Publishing. https://doi.org/10.1787/5jxvl3dwk3f0-en.
Singer, R.S., M.P. Ward, and G. Maldonado. 2006. Can landscape ecology untangle the complexity of antimicrobial resistance? Nature Reviews Microbiology 4: 943–952.
Soonthornchaikul, N., and H. Garelick. 2009. Antimicrobial resistance of campylobacter species isolated from edible bivalve molluscs purchased from Bangkok markets, Thailand. Foodborne Pathogens and Disease 6: 947–951. https://doi.org/10.1089/fpd.2008.0236.
Strategic and Technical Advisory Group on Antimicrobial Resistance (STAG-AMR). Report of the first meeting Geneva, 19–20 September 2013. Available from: http://www.who.int/drugresistance/stag/amr_stag_meetingreport0913.pdf.
Sustainable Control of Parasites in Sheep. Available from: http://www.scops.org.uk/.
Thanner, S., D. Drissner, and F. Walsh. 2016. Antimicrobial resistance in agriculture. MBio 7 (2): e02227–e02215. https://doi.org/10.1128/mBio.02227-15.
Van Boeckel, T.P. 2015. Global Trends In Antimicrobial Use In Food Animals. Proceedings of the National Academy of Science 112 (18): 649–5654. https://doi.org/10.1073/pnas.1503141112.
van Bunnik, B.A.D., and M.E.J. Woolhouse. 2017. Modelling the impact of curtailing antibiotic usage in food animals on antibiotic resistance in humans. Royal Society Open Science 4: 161067. https://doi.org/10.1098/rsos.161067.
Wellington, E.M.H., A.B.A. Boxall, P. Cross, E.J. Feil, W.H. Gaze, P.M. Hawkey, A.S. Johnson-Rollings, D.L. Jones, N.M. Lee, W. Otten, C.M. Thomas, and A.P. Williams. 2013. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infectious Diseases 13: 155–165.
World Health Assembly Resolution 51.9. 1998. Emerging and other communicable diseases: Antimicrobial resistance. In 51st world health assembly. Available from: http://apps.who.int/medicinedocs/index/assoc/s16335e/s16335e.pdf?ua=1.
World Health Organisation. 2001. WHO global strategy for containment of antimicrobial resistance. Available from: http://www.who.int/drugresistance/WHO_Global_Strategy_English.pdf.
———. 2011. Tackling antibiotic resistance from a food safety perspective in Europe. Available from: http://www.euro.who.int/__data/assets/pdf_file/0005/136454/e94889.pdf at page 9.
World Health Organisation List of Critically Important Antimicrobials (CIA). 2017. Available from: https://www.who.int/foodsafety/publications/antimicrobials-sixth/en/
Zhu, Y.-G., T.A. Johnson, J.-Q. Su, M. Qiao, G.-X. Guo, R.D. Stedtfield, S.A. Hashsham, and J.M. Tiedje. 2013. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proceedings of the National Academy of Science 110 (9): 3435–3440.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence and indicate if changes were made. The images or other third party material in this chapter are included in the chapter’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the chapter’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Boden, L., Mellor, D. (2020). Epidemiology and Ethics of Antimicrobial Resistance in Animals. In: Jamrozik, E., Selgelid, M. (eds) Ethics and Drug Resistance: Collective Responsibility for Global Public Health. Public Health Ethics Analysis, vol 5. Springer, Cham. https://doi.org/10.1007/978-3-030-27874-8_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-27874-8_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-27873-1
Online ISBN: 978-3-030-27874-8
eBook Packages: Religion and PhilosophyPhilosophy and Religion (R0)