Abstract
Accurate segmentation of brain tumors is critical for clinical quantitative analysis and decision making for glioblastoma patients. Convolutional neural networks (CNNs) have been widely used for this task. Most of the existing methods integrate the multi-modality information by merging them as multiple channels at the input of the network. However, explicitly exploring the complementary information among different modalities has not been well studied. In fact, radiologists rely heavily on the multi-modality complementary information to manually segment each brain tumor substructure. In this paper, such a mechanism is developed by training the CNNs like the annotation process by radiologists. Besides, a 3D lightweight CNN is proposed to extract brain tumor substructures. The dilated convolutions and residual connections are used to dramatically reduce the parameters without loss of the spatial resolution and the number of parameters is only 0.5M. In the BraTS 2018 segmentation task, experiments with the validation dataset show that the proposed method helps to improve the brain tumor segmentation accuracy compared with the common merging strategy. The mean Dice scores on the validation and testing dataset are (0.743, 0.872, 0.773) and (0.645, 0.812, 0.725) for enhancing tumor core, whole tumor, and tumor core, respectively.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Glioblastoma is the most common primary malignant brain tumor [17]. Medical imaging technologies play an important role in the diagnosis, preoperative planning, intraoperative navigation, and postoperative evaluation of the brain cancer. Magnetic Resonance Imaging (MRI) is the most frequently used imaging method in the clinical routine of brain tumors, because it is noninvasive and free of radiation.
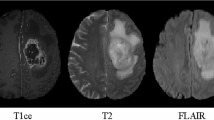
Brain tumor segmentation in multi-modality MRI scans is crucial for the quantitative analysis in clinic. However, it is time-consuming and labor-intensive for radiologists to manually delineate brain tumors. Automatic segmentation of brain tumors in multi-modality MRI scans has a potential to provide a more effective solution, but due to the highly heterogeneous appearance and various shapes of brain tumors, it is one of the most challenging tasks in medical image analysis. Figure 1 presents a brain tumor case and the corresponding label in the BraTS 2018 training dataset.
In recent years, convolutional neural networks (CNNs) have emerged as a powerful tool for medical image segmentation tasks, including organ and lesion segmentation, and achieved unprecedented accuracy. Benefiting from the multimodal brain tumor segmentation challenge [15] which is long-term and competitive, many CNN architectures have been proposed and also achieved state-of-the-art performance. In [13], Kamnitsas et al. constructed an Ensemble of Multiple Models and Architectures (EMMA) for robust brain tumor segmentation including two deepMedic models, three 3D FCNs, and two 3D U-Nets. Wang et al. [19] developed a cascade of fully convolutional neural networks to decompose the multi-class segmentation problem into a sequence of three binary segmentation problems according to the brain tumor substructures hierarchy and proposed anisotropic networks to deal with 3D images as a trade-off among the receptive field, model complexity and memory consumption. The multi-view fusion was used to further reduce noises in the segmentation results. Isensee et al. [12] modified the U-Net to maximize brain tumor segmentation performance. The architecture consisted of a context aggregation pathway which was used to encode increasingly abstract representations of the input and a localization pathway which was designed to transfer the low level features to a higher spatial resolution.
Most of the existing multi-modality brain tumor segmentation methods use an early-fusion strategy which integrates the multi-modality information from the original MRI scans. For example, four MRI modalities (T1, T2, T1ce, and Flair) are simply merged as four channels at the input of the network [12, 13, 19]. However, as argued in [18] in the context of multi-modal learning, it is difficult to discover highly non-linear relationships among the low-level features of different modalities. Besides, early-fusion methods implicitly assume that the relationship among different modalities is simple (e.g., linear) and the importance among these modalities is equal for the segmentation of different brain tumor substructures. In fact, when radiologists manually segment tumor substructures, they pay different attention to different modalities. For example, when segmenting the tumor core, radiologists will pay more attention to T1ce modality rather than Flair or T2 modalities. Thus, the importance of different modalities is not the same when segmenting a specific tumor substructure; The complementary information among these modalities plays an important role to the final brain tumor labels. As far as we know, explicitly exploring the complementary information among different modalities has not been well studied for brain tumor substructures segmentation.
In this paper, we train the networks like the manual segmentation process by radiologists to explicitly explore the complementary information among different MRI modalities. Specifically, the pipeline design of the brain tumor segmentation is guided by clinical brain tumor annotation protocol. In addition, we propose a novel 3D lightweight Convolutional Neural Network (CNN) architecture which captures high-level features from a large receptive field without the loss of resolution of the feature maps. The proposed lightweight CNN makes a good balance between the 3D receptive field and model complexity. It has only ten hidden layers and the number of parameters is only 0.5M. We evaluate the proposed lightweight CNN architecture on the BraTS 2018 validation and testing dataset and achieve the promising segmentation results. Besides, experiments show that an improvement of segmentation accuracy is achieved by exploring the complementary information among different modalities.
2 Methods
2.1 MRI Modality Analysis and Selection of Brain Tumors
The MRI modality selection method is inspired by how radiologists segment the brain tumor substructures. From [15], it can be found that different brain tumor substructures are annotated by different strategies in clinic. Specifically, the edema (belongs to the whole tumor) is segmented primarily from T2 images. Flair images are used to cross-check the extension of the edema. The enhancing tumor and the tumor core are identified from T1ce images. Motivated by this annotation protocol, different modalities are selected for the segmentation of different brain tumor substructures. Table 1 presents an overview of the used modalities for different substructures segmentation. Briefly, like annotation process by radiologists, we mainly use the Flair and the T2 modalities to segment the whole tumor and use the T1ce modality to segment the enhancing tumor and the tumor core.
2.2 Proposed 3D Lightweight CNN Architecture
Although for 3D volume data segmentation, traditional 3D architectures such as 3D U-Net and FCN, have high memory consumption in the training phase, the 3D context information would be degenerated if changing the inputs as 2D or 2.5D slices to relieve the computational burden. As a trade-off between memory consumption and 3D context information, a 3D lightweight CNN architecture (Fig. 2) is proposed for 3D brain tumor segmentation which integrates the dilated convolution with different dilated rates and residual connections. Table 2 presents the detailed configurations of the proposed architecture.
Dilated Convolution with Increasing and Decreasing Dilated Rates. Dilated convolutions have been verified as a very effective structure in deep neural networks [6, 21]. The main idea of dilated convolution is to insert “holes” among pixels in traditional convolutional kernels to enlarge the respective field. In order to obtain multi-scale semantic information, we employ different dilation factors in the proposed architecture. The dilation factors are set to 1, 2, and 4 with the increasing and decreasing sequences which can avoid the gridding effect of the standard dilated convolution [8, 20].
Residual Connections. To train deep CNNs more effectively, residual connections were first introduced by He et al. [10]. The main idea of residual connections is to learn residual functions through the use of identity-based skip connections which ease the flow of information across units. Our proposed lightweight architecture adds residual connections to each dilated convolutional layer. In addition, each convolutional layer is associated with a batch normalization layer [11] and an element-wise parametric rectified linear Unit (prelu) layer [9] to speed up the convergence of the training process.
2.3 Two-Stage Cascaded Framework
Cascaded strategy has been proved to be an effective way for brain tumor substructures segmentation [19] in the BraTS 2017. Inspired by this work, we deal with the task with a two-stage cascaded framework. Figure 3 presents the whole pipeline. The lightweight CNN architecture is iteratively used to sequentially segment brain tumor substructures. In the first stage, the whole tumor is segmented from Flair and T2 modalities. The segmentation results of Flair and T2 modalities are merged by simply making an union to generate a bounding box of the whole tumor region of interest (ROI). Besides, an extension with 5 pixels is applied to the whole tumor ROI bounding box so as to avoid possible under-segmentation. Specifically, each side of the bounding box is relaxed by 5 pixels. The whole tumor segmentation from the Flair modality is used as the final whole tumor segmentation result. Besides, we also try to use the union segmentation from Flair and T2 modalities segmentation results as the final whole tumor segmentation result, but there is no improvement of the accuracy. In the second stage, the corresponding T1ce images in the ROI are used to train a new 3D lightweight CNN to make a triple prediction for the enhancing tumor and the tumor core segmentation.
3 Experiments and Results
3.1 Preprocessing
The proposed method was evaluated on the Brain Tumor Segmentation Challenge (BraTS 2018) dataset [2,3,4]. The training dataset consisted of 210 cases with high grade glioma (HGG), 75 cases with low grade glioma (LGG) and the corresponding manual segmentation. Each case had four 3D MR modalities (T1, T2, Flair, and T1ce).
To enforce the MR volume data to be more uniform, the following preprocessing strategies (Table 3) were applied to the used modalities. It can be seen that the Flair modality is added an additional histogram equalization compared to the preprocessing methods for the remained modalities. This was because the intensity distributions of the Flair images vary considerably across different cases. Figure 4 presents two examples of Flair modalities in the training dataset. Obviously, the intensity distributions of the two cases still differed remarkably after the z-score normalization. Therefore, the histogram equalization was further applied to make them share similar intensity distribution. For the T2 and T1ce modalities, however, there were no such significant intensity differences among different cases, so a simple z-score preprocessing was enough.
Preprocessing results of two Flair images. The first row is the case named “Brats18_TCIA02_135_1 (78th slice)” and the second row is the case named “Brats18_TCIA02_283_1 (78th slice)”. After z-score normalization, there is still a great difference between the two images (the 2nd column). Further, the histogram equalization is applied to make them share similar intensity distributions (the 3rd column).
3.2 Implementation Details
The BraTS 2018 training dataset was randomly divided into training data (80%), validation data (10%), and test data (10%) to find the proper parameters. After that, all the training data were employed to train the final models which were used for the official validation and testing dataset.
The proposed networks were implemented in tensorflow [1] and NiftyNet [7, 14]. The input 3D volume data was resized to \(64*64*64\) by the first order spline interpolation. The predicted segmentation was also resized in the same way to retrieve the original 3D volume. The batch size was set to 2 and the maximum number of iterations was 10k. The optimizer was the adam with an initial learning rate 0.001. The loss function was Dice coefficient [16] which can deal with the data imbalance. A L2 weight decay of \(10^{-5}\) was used. No external data was used and data augmentation included random rotation, random spatial scaling, and random flipping. The whole training process cost about 30 h on a desktop with an Intel Core i7 CPU and a NVIDIA 1080Ti GPU.
Segmentation results of the brain tumor substructures from the BraTS 2018 validation dataset (named “Brats18_CBICA_ALV_1”) by the proposed method (complementary fusion) and its variant (ordinary fusion). Green: edema; red: necrotic and the non-enhancing tumor core; yellow: enhancing tumor core. The obvious mis-segmentations of the non-enhancing tumor core are highlighted by white arrows. (Color figure online)
3.3 Segmentation Results
We test our framework on the BraTS 2018 validation dataset with 66 new cases. To evaluate whether the proposed method (termed as complementary fusion) could improve the brain tumor segmentation results, we compare it with the ordinary strategy which merges four MR modalities as four channels at the input of the network. The whole pipeline is also a two-stage cascaded way and we refer to it as ordinary fusion. Except the difference at the input of the network, all the hyper-parameters of the ordinary fusion are the same with the proposed complementary fusion strategy during the training process.
Table 4 presents quantitative evaluations of the proposed method (complementary fusion) and its variant (complementary fusion) on the BraTS 2018 validation dataset. For the ordinary fusion, the Dice scores are 0.709, 0.851, and 0.751 for enhancing tumor core, whole tumor, and tumor core respectively. For the proposed complementary fusion, an improvement is achieved, and the Dice scores are 0.743, 0.872, and 0.773 for these substructures respectively.
Figure 5 shows an example for the brain tumor substructures segmentation from the BraTS 2018 validation dataset. Three views are presented, including the axial view, the sagittal view, and the coronal view. For the simplicity of visualization, only the Flair and T1ce images are shown, because the two modalities can clearly display the whole tumor, enhancing tumor, and tumor core. The first and the second columns present the input images from Flair and T1ce modalities, respectively. We have compared the proposed method with its variant that employed the ordinary fusion method at the input. The third and the fourth columns show the ordinary fusion and the complementary fusion segmentation results, respectively. The green, red, and yellow colors show the edema, tumor core, and enhancing tumor, respectively. It can be observed that the predictions by the ordinary fusion seem to have an over segmentation (highlighted by white arrows) of the tumor core. When using the complementary fusion, the segmentation results are more accurate.
Table 5 presents quantitative evaluations with the BraTS 2018 testing dataset. It shows the mean values, standard deviations, medians, Dice, and 25 and 75 quantiles of the 95th Hausdorff distance. Compared with the performance on the validation dataset, the performance on the testing dataset is lower, with average Dice scores of 0.645, 0.812, and 0.725 for enhancing tumor core, whole tumor, and tumor core, respectively. The higher median values show that good segmentation results are achieved for most cases, and some outliers lead to the lower average scores. The ranking analysis is reported in [5].
4 Discussion and Conclusion
There are several advantages of the proposed framework. Firstly, the complementary information among different modalities is explicitly explored to segment brain tumor substructures which can avoid the interference from other confusing modalities as well as reducing the complexity compared with using all the modalities as inputs simultaneously. Besides, the proposed 3D lightweight CNN effectively uses the dilated convolutions to enlarge the receptive fields and to aggregate the global information. The increasing and decreasing arrangement of the dilate factors can alleviate the gridding effect caused by the standard dilated convolutions. The architecture is very compact and computation efficient. Finally, the cascaded CNNs, which have been proved to be an effective strategy, can separate the complex multiple class segmentation into simper problems and reduce false positives by spatial constrains of brain tumor anatomical structures.
In conclusion, we explicitly explore the complementary information among different modalities according to the clinical annotation protocol. In addition, a compact 3D lightweight CNN architecture is proposed and the number of parameters is only 0.5M. The proposed approach achieves a promising performance on the BraTS 2018 validation and testing dataset. Experiments with the BraTS 2018 validation dataset show that the complementary fusion strategy helps to improve the brain tumor segmentation accuracy compared with the ordinary fusion method.
References
Abadi, M., et al.: TensorFlow: large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467 (2016)
Bakas, S., et al.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 4, 170117 (2017)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. Cancer Imaging Arch. (2017)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. Cancer Imaging Arch. (2017)
Bakas, S., Reyes, M., Menze, B., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv preprint arXiv:1811.02629 (2018)
Chen, L., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 40(4), 834–848 (2018)
Gibson, E., et al.: NiftyNet: a deep-learning platform for medical imaging. Comput. Methods Programs Biomed. 158, 113–122 (2018)
Hamaguchi, R., Fujita, A., Nemoto, K., Imaizumi, T., Hikosaka, S.: Effective use of dilated convolutions for segmenting small object instances in remote sensing imagery. In: 2018 IEEE Winter Conference on Applications of Computer Vision, WACV 2018, Lake Tahoe, NV, USA, 12–15 March 2018, pp. 1442–1450 (2018)
He, K., Zhang, X., Ren, S., Sun, J.: Delving deep into rectifiers: surpassing human-level performance on imagenet classification. In: 2015 IEEE International Conference on Computer Vision, ICCV 2015, Santiago, Chile, 7–13 December 2015, pp. 1026–1034 (2015)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
He, K., Zhang, X., Ren, S., Sun, J.: Identity mappings in deep residual networks. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9908, pp. 630–645. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46493-0_38
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: Brain tumor segmentation and radiomics survival prediction: contribution to the BRATS 2017 challenge. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 287–297. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_25
Kamnitsas, K., et al.: Ensembles of multiple models and architectures for robust brain tumour segmentation. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 450–462. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_38
Li, W., Wang, G., Fidon, L., Ourselin, S., Cardoso, M.J., Vercauteren, T.: On the compactness, efficiency, and representation of 3D convolutional networks: brain parcellation as a pretext task. In: Niethammer, M., et al. (eds.) IPMI 2017. LNCS, vol. 10265, pp. 348–360. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-59050-9_28
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015)
Milletari, F., Navab, N., Ahmadi, S.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: Fourth International Conference on 3D Vision, 3DV 2016, Stanford, CA, USA, 25–28 October 2016, pp. 565–571 (2016)
Ostrom, Q.T., et al.: CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the united states in 2008–2012. Neuro-oncology 17(suppl-4), iv1–iv62 (2015)
Srivastava, N., Salakhutdinov, R.: Multimodal learning with deep boltzmann machines. J. Mach. Learn. Res. 15(1), 2949–2980 (2014)
Wang, G., Li, W., Ourselin, S., Vercauteren, T.: Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 178–190. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_16
Wang, P., et al.: Understanding convolution for semantic segmentation. In: 2018 IEEE Winter Conference on Applications of Computer Vision, WACV 2018, Lake Tahoe, NV, USA, 12–15 March 2018, pp. 1451–1460 (2018)
Yu, F., Koltun, V.: Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv:1511.07122 (2015)
Acknowledgements
This work is supported by National Nature Science Foundation of China (No: 11531005). And we also would like to thank the NiftyNet team, they developed the open source convolutional neural networks platform for medical image analysis, which made us more efficiently to build our model. Last but not least, we gratefully thank the BraTS organizers and data contributors for their efforts on hosting the excellent challenge.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Ma, J., Yang, X. (2019). Automatic Brain Tumor Segmentation by Exploring the Multi-modality Complementary Information and Cascaded 3D Lightweight CNNs. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science(), vol 11384. Springer, Cham. https://doi.org/10.1007/978-3-030-11726-9_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-11726-9_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-11725-2
Online ISBN: 978-3-030-11726-9
eBook Packages: Computer ScienceComputer Science (R0)