Abstract
Stroke is an important neuro-vascular disease, for which distinguishing necrotic from salvageable brain tissue is a useful, albeit challenging task. In light of the Ischemic Stroke Lesion Segmentation challenge (ISLES) of 2018 we propose a deep learning-based method to automatically segment necrotic brain tissue at the time of acute imaging based on CT perfusion (CTP) imaging. The proposed convolutional neural network (CNN) makes a voxelwise segmentation of the core lesion. In order to predict the tissue status in one voxel it processes CTP information from the surrounding spatial context from both this voxel and from a corresponding voxel at the contra-lateral side of the brain. The contra-lateral CTP information is obtained by registering the reflection w.r.t. a sagittal plane through the geometric center. Preprocessed training data was augmented during training and a five-fold cross-validation was used to experiment for the optimal hyperparameters. We used weighted binary cross-entropy and re-calibrated the probabilities upon prediction. The final segmentations were obtained by thresholding the probabilities at 0.50 from the model that performed best w.r.t. the Dice score during training. The proposed method achieves an average validation Dice score of 0.45. Our method slightly underperformed on the ISLES 2018 challenge test dataset with the average Dice score dropping to 0.38.
J. Bertels is part of NEXIS, a project that has received funding from the European Union’s Horizon 2020 Research and Innovations Programme (Grant Agreement #780026).
D. Robben is supported by an innovation mandate of Flanders Innovation & Entrepreneurship (VLAIO).
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Stroke is the main cause of neurological disability in older adults [2]. Up to four out of five acute stroke cases is ischemic. In these cases, the ischemia is the result of a sudden occlusion of a cerebral artery. If a large artery is occluded, there is a possibility that mechanical thrombectomy (i.e. the intra-arterial removal of the cloth) can improve patient outcome [4]. Important biomarkers for the correct selection of patients are the volumes of necrotic and salvageable tissue, respectively the core and penumbra. In that light, this year’s Ischemic Stroke Lesion Segmentation (ISLES) challenge [1] asked for methods that can automatically segment core tissue based on CT (perfusion) imaging.
Both the training data (blue) and test data (red) are heterogeneous w.r.t. certain image properties. The y-axis shows the number of data samples (i.e. slabs). Top left: The in-plane resolution ranges from 0.785 to 1.086 mm. Top right: The slice thickness ranges from 4.0 to 12.0 mm. Bottom left: The number of slices ranges from 2 to 22. Bottom right: The number of time points (at the initial resolution of 1 image/1 s) ranges from 28 to 64. (Color figure online)
With respect to the detection of core tissue, a standard CT is unable to capture early necrotic changes and will therefor result in underestimation [5]. A more sensitive approach is to acquire a perfusion scan and extract certain perfusion parameters of the parenchymal tissue via deconvolution analysis [3]. As such, core tissue is characterized with an increase in Tmax and MTT, and a decrease in CBF and CBV. During last year’s ISLES challenge it was shown that convolutional neural networks (CNNs) can be used to predict the final infarction based on these parameter maps (and additional patient and treatment information). In parallel, it has been found that CNNs can be used to estimate the perfusion parameters directly, hence bypassing the mathematically ill-posed deconvolution problem [7]. Nonetheless, these CNN-based methods still require an arterial input function (AIF) to do the deconvolution.
This work makes a contribution on two levels. First, we are the first to strictly limit the CNN to use only CT perfusion (CTP) data as input in the prediction of core tissue. This holds both during training and testing. Second, we explore the alternative use of contra-lateral information instead of the explicit manual or (semi-)automatic selection of the AIF. We identify the ISLES 2018 challenge as the perfect setup to compare the performance directly with other state-of-the-art methods. In the next section we briefly introduce the dataset and some initial preprocessing. In Sect. 3, we highlight the extraction of contra-lateral information and the experimental setup, including the architecture of our CNN. In Sect. 4, we show the validation results on the ISLES 2018 training set as well as the results on the test set.
2 Data
The ISLES 2018 dataset consists of acute CT and CTP images, and the derived Tmax, CBF and CBV perfusion maps, from 125 patients. The ground truth cores were delineated on the MR DWI images, which were acquired soon thereafter. The participants only have access to the training dataset. For some patients there are two slabs to cover the lesion. We consider each slab independently, resulting in 94 slabs present in the training dataset and 62 in the test dataset.
The in-plane resolutions are isotropic and we notice resolutions ranging from 0.796 to 1.039 mm in the training set and from 0.785 to 1.086 mm in the test set (Fig. 1). We consider these spatial resolutions similar enough and avoid resampling the images. The spatial resolution in the axial direction ranges from 4 to 12 mm and the number of slices from 2 to 22. We therefore opt to work in 2D. We further notice the discrepancy in the available number of time points, ranging from 43 to 64 and from 28 to 64, both at a temporal resolution of 1 image/1 s, for training and test sets respectively. We first resample the signal along the time axis to a temporal resolution of 1 image/2 s by using a smoothing kernel of [1/4, 2/4, 1/4] with a stride of 2. We then pad the signal by repeating the final value until we have 32 time points (i.e. 64 s). The resulting volumes have lower temporal noise and identical shape.
3 Method
As we hypothesised in Sect. 1, we will investigate whether CTP information only, but complemented with contra-lateral information, can be used to predict the core lesion. We will use DeepVoxNet [6] as a framework for doing segment-based training and extensive parallel data augmentation on the CPU. First, we explain how to construct the input for our CNN. Then, the architecture of our CNN will be discussed. Finally, we detail some further training methodologies.
3.1 Contra-Lateral Information
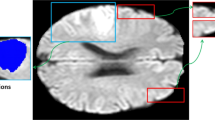
Because an ischemic stroke typically occurs uni-laterally, we want to enrich the information in one voxel (i.e. currently a time series of 32 points depicting the passage of contrast) with the information present in a corresponding voxel at the contra-lateral side of the brain (Fig. 2). This way each voxel contains information from at least one healthy voxel, thus how the perfusion or contrast passage could look like in similar but healthy parenchyma. For this purpose, we average each CTP over time and clip values in the range of [0, 100] HU. We then flip the image laterally (i.e. w.r.t. a sagittal plane) across its geometrical center and rotate the flipped image back to minimize the mean squared error (MSE). This rotation is applied to the unclipped, flipped volume and the result is concatenated to the original CTP volume. Each voxel now has 64 features. Before we let the network crunch this data, we make sure to suppress the influence of extreme outliers (e.g. streak artifacts) via clipping in the range of [0, 150] HU. We furthermore normalize the data to zero mean and unit variance.
In order to complement each voxel with contra-lateral information, we perform the following operations. Left: We flip the entire original volume (blue) w.r.t. a sagittal axis through the geometrical center (yellow) to obtain the contra-lateral volume (red). Middle: We rotate the clipped (in [0, 100] HU) contra-lateral volume for the lowest mean squared error (MSE). Right: We apply this angle (here 0.28 radians for case 4 of the training dataset) to the contra-lateral, unclipped volume and concatenate with the original to obtain 64 features for each voxel. (Color figure online)
3.2 Data Augmentation
To artificially increase the number of training samples and regularize the network as such we augment input images on the level of the image and on the level of the segments, which are extracted from the image. At the image level, the image is rotated with 0.5 probability according to \(\mathcal {N}\)(0, 10\(^{\circ }\)) and Gaussian noise \(\mathcal {N}\)(0, 0.03) is added to the original and flipped part independently. At the segment level, the segments can be flipped laterally with 0.5 probability, the intensities can be shifted \(\mathcal {N}\)(0, 0.01) and scaled \(\mathcal {N}\)(1, 0.01), and the time series can be shifted forward or backward in time \(\mathcal {U}\)(\(-6\), 6). We also perform contrast scaling of the segments, where we scale the intensity differences of a certain time point w.r.t. the first time point according to a Lognormal distribution with zero mean and a standard deviation of 0.3 [5].
3.3 Class Balancing
Based on the images obtained before normalization we construct a binary head mask from where the intensities lie in the range [0, 150] HU. This head mask is used to mask the output of the network both during training and testing. Network input segment centers are sampled uniformly from within this region. In order to further balance the class observations we use a weighted binary cross-entropy with weights calculated to equalize the prior class probabilities across the training data. Upon prediction we re-calibrate the probabilities.
The CNN takes as input a \(97\times 97\) 2D segment with 64 features, sampled from the concatenated original and contra-lateral volumes (see Fig. 2) and outputs lesion probabilities for the corresponding \(29\,\times \,29\) segment. Our U-Net-like architecture consists of two pooling (green arrows) and two up-sampling layers (red arrows). Together with all the \(3\times 3\) convolution layers (blue layers; each of which has 64 filters) involved, this results in a receptive field of \(69\times 69\) voxels. The final three convolution layers are fully-convolutional with 128 filters. (Color figure online)
3.4 CNN Architecture
We use a U-Net-like [8] architecture with two pooling and two up-sampling layers, which respectively downsample or upsample by a factor of three (Fig. 3). Before each pooling or up-sampling layer (and after the final one), we perform two consecutive convolutions with 64 filters of size \(3\times 3\). Two fully-convolutional layers precede the final layer, each with 128 filters. The parametric ReLU (pReLU) is used as the non-linear activation function. We use batch normalization and local skip connections to improve learning. The CNN is characterized with valid padding, an in-plane receptive field size of \(69\times 69\) voxels and a total number of 470,657 trainable parameters. Both during training and testing we use input segments with a fixed size of \(97\times 97\) with 64 features to predict an output segment of size \(29\times 29\).
3.5 Experiments
We train the CNN using five-fold cross validation on the ISLES 2018 training set. The predictions of each model will be evaluated and averaged during test time. We use the ADAM optimizer for 4000 epochs with an initial learning rate of 10\(^{-4}\) and decrease the learning rate with a factor of 10 after each 1000 epochs. Both decay rate parameters for ADAM were fixed at 0.9. Here, one epoch iterates through 1280 segments, extracted from 16 subjects and processed in batches of 64 segments. We use L1 and L2 weight regularization with weights of 10\(^{-4}\) and 10\(^{-2}\), respectively. Every 20 epochs we run the model on the (left-out) validation set. The model that optimizes the validation cross-entropy and the model that optimizes the validation Dice score, both at a threshold of 0.50, were stored, CE\(^{0.50}\) and D\(^{0.50}\) respectively.
4 Results and Conclusion
We will compare the results of the models that were found optimal during training w.r.t. binary cross-entropy (CE\(^{0.50}\)) and Dice score (D\(^{0.50}\)), and of their optimal threshold derivatives w.r.t. the Dice score, CE\(^{0.11}\) and D\(^{0.30}\) respectively, by varying the segmentation threshold. In Table 1 these results are listed, as well as the result of model D\(^{0.50}\) on the test set (D\(^{0.50}\) @ test). Without post Dice score threshold optimization we notice the better results for the D\(^{0.50}\) model compared to the CE\(^{0.50}\) model. Especially the Dice score and the average volume difference stand out. Optimizing the thresholds for each of those models w.r.t. the Dice score could further improve the performance on this metric. Especially the CE\(^{0.50}\) model benefits from this, explained by the discrepancy between its precision and recall. Although other methods performed better w.r.t. Dice score, we opted for model D\(^{0.50}\) for participating the challenge because of lower surface distances and the lower average volume difference. In Fig. 4 one example segmentation with a Dice score of 0.68 on the validation dataset for our submitted model is depicted.
Although results are promising, further research is needed. For example, the registration of the contra-lateral information is far from ideal. The immediate concatenation could hinder the network to learn the correct use of this contra-lateral information.
References
Ischemic Stroke Lesion Segmentation (ISLES). www.isles-challenge.org
Berkhemer, O.A., et al.: A randomized trial of intraarterial treatment for acute ischemic stroke. N. Engl. J. Med. 372(1), 11–20 (2015). https://doi.org/10.1056/NEJMoa1411587
Fieselmann, A., Kowarschik, M., Ganguly, A., Hornegger, J., Fahrig, R.: Deconvolution-based CT and MR brain perfusion measurement: theoretical model revisited and practical implementation details. Int. J. Biomed. Imaging 2011 (2011). https://doi.org/10.1155/2011/467563
Goyal, M., et al.: Endovascular thrombectomy after large-vessel ischaemic stroke: a meta-analysis of individual patient data from five randomised trials. Lancet 387, 1723–1731 (2016). https://doi.org/10.1016/S0140-6736(16)00163-X
von Kummer, R., Dzialowski, I.: Imaging of cerebral ischemic edema and neuronal death. Neuroradiology 59(6), 545–553 (2017). https://doi.org/10.1007/s00234-017-1847-6
Robben, D., Bertels, J., Willems, S.: DeepVoxNet. Technical report, KU Leuven: KUL/ESAT/PSI/1801 (2018)
Robben, D., Suetens, P.: Perfusion parameter estimation using neural networks and data augmentation. MICCAI SWITCH (2018). https://arxiv.org/abs/1810.04898
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Bertels, J., Robben, D., Vandermeulen, D., Suetens, P. (2019). Contra-Lateral Information CNN for Core Lesion Segmentation Based on Native CTP in Acute Stroke. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science(), vol 11383. Springer, Cham. https://doi.org/10.1007/978-3-030-11723-8_26
Download citation
DOI: https://doi.org/10.1007/978-3-030-11723-8_26
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-11722-1
Online ISBN: 978-3-030-11723-8
eBook Packages: Computer ScienceComputer Science (R0)