Abstract
This chapter presents the contribution of databanks to the development of biodiversity knowledge through the example of marine biodiversity databanks. Focusing on the marine field allows us to insist on the imbalance of the unknown vs. the better known part. The chapter emphasizes the role of taxonomic and genetic databanks as well as the ongoing transformations that databanks are submitted to in order to answer pressing demands due to the biodiversity crisis. It aims to analyse the requirements biodiversity databanks have to satisfy in order to help both researchers and conservationists in their respective endeavors. It begins by pointing out the main characteristics and limits of biodiversity knowledge and defend the view that databanks are well-suited to overcome these limits as soon as they are widely accessible and interoperable. These constraints are analysed as both technical and scientific. Their dynamic dimension is emphasized as databanks must comply with the rapid evolution of scientific knowledge. We also propose a view on the relationships between biodiversity knowledge, assessment, and conservation.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Assessing biodiversity, according to the comparison developed in the Introduction of this volume, is like assessing the state of a patient, that is, trying to infer how bad she is from observations. The double aim of this operation is to guess how her state will evolve in the future and to design actions to improve it. Assessment is thus future- and action-oriented. It benefits from available scientific knowledge and may in turn contribute to its development, but assessment and knowledge do not evolve at the same pace as assessment is submitted to the pressure of time. The relationship between assessment of biodiversity in a given geographical area and scientific knowledge of biodiversity, as built up in evolutionary studies, taxonomy, phylogenetics, population genetics, and ecology, is unbalanced since the corpus that has been scientifically established is usually far from sufficient for the assessment task. The diversity of living beings, of their behaviours and interactions is so huge that what is known about it may be compared to ancient maps in which the size of the known world is much smaller than the breath of the unknown world. Thus, in most cases, currently available knowledge of biodiversity cannot provide but a small contribution to the assessment of biodiversity. However, some parts of this knowledge are more readily useful for assessment tasks than others: the knowledge of biodiversity that is contained in databanks can be easily harnessed. But to what extent can the data in databanks be treated as knowledge? We discuss this question in the following, and emphasize again that retrieving this knowledge cannot be anything else than a small part of the assessment task.
Learning about biodiversity is a complex endeavor. First, because biodiversity itself is a complex object of knowledge. Second, because it extends on virtually every region of our planet, however small. Third, biodiversity knowledge comes from heterogeneous sources: taxonomic, evolutionary (including phylogenetic), genetic, and ecological research. This results in a confused picture in need of clarification. However, in the last few decades, biodiversity knowledge has immensely benefited from the establishment of international databanks. They play a fundamental role in the improvement of the current, confused picture of biodiversity because they provide scientists with elements from which they can develop knowledge of biodiversity at a the global scale.
Our aim in this paper is to examine how biodiversity databanks contribute to developing current knowledge of biodiversity. We do so by putting forward an epistemological analysis of the structure and functioning of databanks, both at the individual and collective (network) level. The epistemological analysis of scientific databanks has greatly benefited from Leonelli’s work (2010, 2013a, b, 2016). Whereas she focused on various fields within biology, she never addressed the topic of biodiversity databanks. With this chapter, we wish to fill this gap and participate in her effort to put databanks in their right place in contemporary biological science and assessment and conservation policies.
The chapter is focused on marine biodiversity databanks and their role in the development of knowledge and assessment of biodiversity. Marine biodiversity is even less known than terrestrial biodiversity, thus illustrating the “ancient map flavour” of this domain of scientific knowledge. By describing how marine biodiversity databanks are developed, we show what kind of knowledge they promote and how it may be used in assessment and conservation tasks.
We begin by analyzing what it means to know about biodiversity. This question is raised because biodiversity is an unusual object of knowledge, crossing spatial and temporal scales. It is thus important to explore how a reasonably unified picture of biodiversity can be achieved by combining various components. Databanks contribute to this picture in an important way. We try to uncover which complex processes result in the “data” that are included in databanks and available for assessment and conservation tasks. We show that these data, far from being “brute data”, are pieces of scientific knowledge subject to constant revision. The second part of the paper is devoted to the current uses of biodiversity databanks and associated requirements for databank designers. Finally, we put forward insights about how to build up biodiversity databanks that could improve our current knowledge of biodiversity.
2 What Does It Mean and What Does It Take to Know Biodiversity?
Because biodiversity knowledge involves evolutionary, taxonomic, and ecological research, as well as attempts at unifying insights from these three domains, it has to face a major conceptual and theoretical challenge. In recent times, this epistemic task has been shaped by a major external factor: urgency. Biologists can no more consider themselves as free of taking their time: they have to hurry up because of the severe crisis biodiversity is currently suffering (Western 1992; Grehan 1993; Takacs 1996; Olson et al. 2002; Singh 2002). The urgency of designing conservation policies induces an acceleration of assessment endeavours, which themselves increase the pressure on knowledge-building.
In this part, we examine in what sense the biodiversity crisis shapes the way we conceive of biodiversity knowledge and of biodiversity itself as an object of scientific knowledge. We first present the main features of the knowledge of biodiversity in its current available form and the difficulties it faces (3.2.1). We then review how it may be improved by the development of appropriate cyber-infrastructures, by examining the question: What are data in biodiversity databanks? (3.2.2) and then by giving examples of cyber-infrastructures (3.2.3).
2.1 Our Current Knowledge of Biodiversity and the Difficulties It Faces
The aim of this section is to describe the main features of the current knowledge of biodiversity. We do so by focusing on the position of taxonomic knowledge within biodiversity knowledge because the slow pace of its development is a major hindrance of assessment and conservation endeavours. We focus on the following tension: On the one hand, many attempts at biodiversity assessment try to circumvent the delays of taxonomic identification, but on the other, taxonomic knowledge appears as an indispensable component of biodiversity knowledge. We complete our description of the current state of biodiversity knowledge and the difficulties it faces by emphasizing how heterogeneous and patchy it is.
From the point of view of taxonomists, taxonomic knowledge, namely, the association of organisms with species names (or at least with genus names, and maybe with variety names), is an indispensable component of biodiversity knowledge and assessment as the descriptions of most components and processes of biodiversity rely on species identification. For sure, some studies are not taxonomy-dependent, like measurements of mass or energy transfer during biological cycles, but taxonomy appears as a main gateway to understanding what is going on at the various spatial and temporal scales where biological processes take place. As such, taxonomic knowledge may be seen to serve as the ground on which other epistemic endeavours within biodiversity studies, including conservation biology, can flourish. To what extent should the taxonomists’ point of view be taken into consideration? In order to answer this question, we present and discuss an example illustrating the position of taxonomic knowledge within biodiversity knowledge: the earth worm example.
Earth worms are well-known, and for long, because they are ecologically important; however, their phylogeny and taxonomic status has long been unclear. Earth worms have been briefly described by Linnaeus in the eighteenth century. He gave them the species name Lumbricus terrestris. At the beginning of the nineteenth century, Savigny put forward a taxonomic revision based on the study of morphological characters (this story is told in James et al. 2010). He hypothesized that the organisms that Linnaeus had called Lumbricus terrestris actually form two species and introduced the name Enterion herculeus to designate the newly recognized species. In 1900, Savigny’s morphological data have been re-interpreted and his revision rejected (James et al. 2010). At that time, the difference between the two sets of characters that Savigny relied upon in favor of taxonomic revision was interpreted as intra-specific polymorphism. However, this was not the end of the story: in 2009, genetic analysis by Richard et al. (2009) detected two homogeneous genetic groups within the set of organisms called Lumbricus terrestris. This led James and co-authors (2010) to begin a new, systematic study that included 230 fresh specimens from Europe and North America (belonging to L. terrestris and other species in the Lombricus genus) and specimens that had been preserved by Savigny. This new study was both genetic and morphological; it took part in the Barcoding earthworms programme and its results have been integrated in BoLD and GenBank, which are two major international, genetic databanks (see below for more details about these databanks). James et al. showed that Savigny was right: there are two diverging groups within L. terrestris. A new revision, similar to the one put forward by Savigny, thus occurred. We are now left with two species of earth worms: Lumbricus terrestris and Enterion herculeus.
What has been the upshot of this history of successive taxonomic revisions? It is strikingly different within and outside taxonomy. Within taxonomy, the earthworm episode is just another example showing that taxonomic knowledge, as all empirical knowledge, is of hypothetical nature and is thus susceptible of being criticized and revised as new data are available. For the specialists of the Lumbricus genus, the state of knowledge has been upgraded in such a way that there are now two well-established species where there used to be only one. Outside taxonomy, the situation is utterly different. Few biologists have realized that the state of taxonomic knowledge has changed within the Lumbricus genus.Footnote 1 The main reason why this is so is worth emphasizing: it is that non-taxonomists do not consider taxonomic knowledge as hypothetical, but rather as established once and for all, which is obviously erroneous. This ignorance has negative consequences: they may believe they have reached firm, well-established results for Lumbricus terrestris whereas they were actually studying Enterion herculeus. Unless they preserved (parts of) the specimens from which they have extracted genetic or physiological materiel, it is impossible to know what species they are talking about in their publications, L. terrestris or Enterion herculeus. As a result, their studies are simply pointless as they cannot possible lead to any useful conclusion. Imagine the same error about a species of mosquitos—the economical and medical consequences would be huge.
The lesson we may draw from the earth worm example is that taxonomic knowledge cannot be ignored by non-taxonomists. When they happen to use outdated taxonomic knowledge, their results are threatened. In some cases, the impact of taxonomic revisions may only bear on taxonomy itself, but in others, the chain of consequences may affect other parts of biology, including conservation biology and biodiversity assessment. Thus, with respect to taxonomy, biodiversity knowledge is not what it should be, as non-taxonomists do not use it appropriately. This appears as a major difficulty facing the development of biodiversity knowledge. It is not the only one. In the remaining of this section, we briefly discuss two other features of biodiversity knowledge that forbid it to provide us with a clear and unified picture of biodiversity, namely its heterogeneity and patchiness.
As it is built up from elements coming from various origins (taxonomy, phylogenetic studies, ecology, macro-ecology, biogeography, and evolution studies), biodiversity knowledge is dis-unified in such a way that it is unable to provide people in charge of assessment or conservation policies with any kind of firm ground. The main reason for this lack of unity is that each involved discipline has its own units, which are difficult to compare with one another. For instance, in population genetics, a gene may be considered a unit of biodiversity (but as is well-known, a gene in population genetics is not exactly the same entity as a gene in molecular biology and genetic databanks, which renders things even more complicated (Baetu 2012; Carlson 1991; Falk 1986; Fogle 2000; Gerstein et al. 2007; Kitcher 1982; Sterelny and Kitcher 1988; Waters 1994). In conservation biology, it is not uncommon to count organisms as units of biodiversity. Populations, species, communities, ecosystems, or even landscapes are also relevant units of biodiversity in ecology and conservation biology. But how do these different units compare? There is no general theory yet that would be able to precisely connect genes with organisms, or a fortiori with communities or ecosystems. With respect to spatial units, things are not better, as fragments of millimeters are as important as regional scales or even the whole surface of the Earth. The same is true with temporal units. Even though, from the theoretical point of view, all the involved disciplines are somehow unified by the theory of evolution, in practice, unification faces many obstacles due to the diversity of relevant units.
Besides being heterogeneous, the knowledge of biodiversity is also patchy in many ways. First, the conceptual links among taxonomy, phylogenetic studies, ecology, macro-ecology, biogeography, and evolution studies are not strong enough to provide biodiversity knowledge with firm theoretical structure (see for instance Leonelli 2009; Sarkar 2016). For instance, the relationships between ecology and evolution studies are notoriously difficult to assess,Footnote 2 besides other difficulties, like differences in temporal scales. Second, a bunch of other difficulties affect the development of biodiversity knowledge, like the large diversity of involved spatial and temporal scales, the difficulty to access certain zones, like deep sea, and the crude fact that certain taxonomic groups, like tunicates (marine invertebrates, sub-group of the Chordates), are much less knownFootnote 3 that fish or crustacean decapods (Bouchet 2006).
The upshot of all this is that our knowledge of biodiversity, in its current state, is unable to provide biodiversity assessments and conservation policies with the firm epistemic ground that they might hope for. Besides being hindered by the awkward position of taxonomy among the other disciplines of biology, biodiversity knowledge is heterogeneous and patchy whereas it should be as unified as possible because biodiversity is an object of knowledge that extends over the whole planet instead of being the object of a series of local, disconnected pieces of knowledge. The latter is testified by the very foundation of GBIF, the Global Biodiversity Information Facility, as described below. The existence of these difficulties forces biologists and conservationists to work at strategies of improvement. In the next section, we present how such improvement may be achieved.
2.2 Improving Our Knowledge of Biodiversity via Cyber-Infrastructures
In order to improve our knowledge of biodiversity, it is first necessary to establish what its vehicles are. By “vehicles”, we mean the devices that are used by researchers to acquire and develop biodiversity knowledge. Among these vehicles, scientific papers play a major role, but they are far from being the only way to build up and complement biodiversity knowledge. Expert reports and outcomes of inventories and assessment tasks are also among the means that researchers can rely on, as well as gene sequences and specimens in natural history collections.
Inventories, gene sequencing, collective scientific expertise, results of assessment endeavours, and collection management are key vehicles of biodiversity knowledge. As most of their outcomes are made available as “data” within databanks, databanks are indispensable vehicles of biodiversity knowledge as well. The “data” they contain are immensely diverse, from gene sequences to species descriptions, taxonomic revisions, geographical localisations, etc. “Data” is thus an ambiguous term that has to be further analyzed. We do so at the end of this section. In the mean time, we put forward a brief history of biodiversity databanks, which is part of the history of cyber-infrastructures Bastow and Leonelli (2010) have analyzed. These authors rightly emphasize that “databases and other online resources have become a central tool for biological research”. Hereby, we present some historical elements relative to biodiversity databanks and emphasize their specific international features.
2.2.1 A Brief History of Biodiversity Databanks
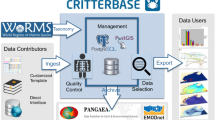
Less than 10 years after the ratification of the Convention on Biological Diversity (CBD),Footnote 4 several databanks were created, whose main objective was to collect geographic and taxonomic data. Among these, we will focus our discussion on the ones relative to the marine field. They are the result of intense collective work during the 1990s aiming at the standardization and organization of data types. We present the development of this collective work in Fig. 3.1. Figure 3.2 shows its results.
The very first databanks have been devoted to taxonomical classification, like the Integrated Taxonomic Information System (ITIS), itself derived from the National Oceanographic Data Center, a former databank from US NOAA (National Oceanographic and Atmospheric Administration). The Taxonomic Database Working Group (TDWG), which first worked on plants, then had an important role in the elaboration of data standards, as well as the bioinformatics working group called “Global Initiative Taxonomy” (OECD 1999). The Global Initiative Taxonomy was the first step toward the creation of the GBIF databank, a few years later (Wieczorek et al. 2012).
Let us now review some outputs of the collective work aimed at the constitution of biodiversity databanks. First, it must be emphasized that the taxonomic impediment (here presented in all its urgent details: https://www.cbd.int/gti/problem.shtml) is a crucial problem facing the development of biodiversity databanks: taxonomists are too few and too slow to cope with the urgency of the biodiversity crisis and do not manage to catch up with extinction rates, thus leaving many extinct species unnamed and un-described. In the 1990s, in the same period in which standardization of biodiversity data was occurring, Bionet International (https://www.uia.org/s/or/en/1100052951), a technical cooperative network for taxonomy, was launched in order to foster exchanges of taxonomic knowledge among different countries and help facing the taxonomic impediment. This was the pre-condition for developing unified knowledge by trying to overcome the heterogeneity of taxonomic expertise among countries.
Against this historical background, it is important to emphasize that biodiversity knowledge relies on the development of genetic as well as taxonomy-based databanks. The most important genetic databank is GenBank (see below). It is commonly used as a resource for biodiversity knowledge and assessment even though its primary goal is not biodiversity-oriented.
By contrast, BoLD (Barcoding of Life Databank) focuses on the link between genetic sequences and taxonomic practices. It aims at being part of a genetic tool for quick species identification. BoLD, and its marine component MarBol, complement traditional taxonomy-oriented databanks like Ocean Biogeographic Information System (OBIS, http://www.iobis.org/), World Register of Marine Species (WoRMS, http://www.marinespecies.org/), and Encyclopedia of Life (EOL, http://eol.org/), which is not restricted to marine organisms. These databanks have been fueled by data gathered during the Census of Marine Life project (CoML 2000–2010) aimed at collecting data and providing researchers with bioinformatics tools. This was an international project gathering 2700 scientists who collected taxonomic, geographic, and ecological data from 540 oceanographic expeditions.
At the beginning, databanks focused on taxonomic classification and upgrading thereof, but they soon had to face a new challenge: connecting taxonomic data with data coming from other kinds of classifications, like genetic, biological and ecological classifications. For instance, biological classifications deliver data like attributes of life stages, reproduction, body size, behavior, feeding method, and diet (Costello et al. 2015). Establishing easy-to-retrieve connections among taxonomic data and biological or ecological traits is an important means to providing conservation biologists with indicators of the not-well-being of ecosystems (e.g., impact of pollution, of fishery, of climate change). To do so, some traits in ecosystems (like reproduction rates and features of habitats) must be described and named on a standardized basis. This cannot be achieved unless a robust consensus has been reached within the scientific community of both ecologists and taxonomists. Marine Species Traits (http://www.marinespecies.org/traits/) aims at the generation of these traits from taxonomic (WoRMS) and geographic databanks (OBIS), which requires and important work of coordination and terminological standardization, as emphasized by Costello et al. (2015). The latter recall that “a rich terminology surrounds descriptions of a species biology and ecology, with sometimes different definitions for the same terms, synonymous terms, and context dependent (e.g., habitat) terms. This terminology has developed over several hundred years of natural history, in different languages, and often terms have multiple meanings in common use.” This requires databanks’ designers to perform scientifically-informed terminological regimentation. The example of Marine Species Traits illustrates how biodiversity databanks, first developed by taxonomists, then connected with genetic databanks, now tend to diversify in order to account for other aspects of biodiversity. This tendency is however difficult to implement because elements of knowledge are much less standardized in ecology than they are in taxonomy and in molecular genetics.
2.2.2 Biodiversity Cyber-Infrastructures
How do the biodiversity databanks we have mentioned so far play contribute to the development of biodiversity knowledge? First, they make data easily accessible. Second, they allow for data being interoperable in a sense to be discussed below. More generally, they organize the vast amounts of data that are relevant to biodiversity study. We discuss and illustrate this three aspects in the following.
Let us begin with accessibility of data. As already emphasized by Leonelli (2010), accessibility of data through internet-based databanks is a major pre-condition for knowledge-building. However, common accessibility, namely, the capacity of a piece of data to be easily retrieved at any scientific institution in the world, without overcoming outrageous paywalls, is not enough to define a useful biodiversity databank. Common accessibility is just the baseline condition of any useful scientific databank. We define scientific “usefulness” in this context as the capacity of databanks to facilitate and accelerate the work of researchers and increase the validity of their results. This cannot only be done by gathering data; organizing data of various origins and nature is instrumental. This involves links among, not only within databanks. Let us emphasize what organizing data and data flows means. Biodiversity databanks, considered collectively, are not as efficient as they could be when information is scattered and when data come from heterogeneous origins. A good biodiversity databank thus should provide researchers with a unique entry to a variety of types of data, e.g., genetic and taxonomic data from various geographical zones. We may illustrate this point with sampling-event data (https://www.gbif.org/sampling-event-data). They report the presence of an organism of such or such species (usually rare or endangered, but not always) together with its spatiotemporal location. These taxonomic, geographical, and temporal data may be provided by amateurs or professional taxonomists. Multiple programmes contribute to produce sampling-event data but they do so for different purposes, some of which might be related with conservation efforts, others with the development of ecological models or the study of migrations and the effects of climate change. This heterogeneity of purpose may generate confusion as to how these data should be stored and used. A scientifically useful sampling-event databank (in our sense) should organize access to information and information flow in such a way as to diminish heterogeneity of origin and facilitate integration of data into more structured pieces of knowledge.
We now turn to interoperability, which can be defined as “the ability of two or more systems or components to exchange information and to use the information that has been exchanged” (Covitz 2004, quoted by Leonelli 2013a). The increased number of databanks involved the ratification of data standards in order to facilitate interoperability among databanks. This allows any individual databank to function as data provider for other databanks, thus facilitating accessibility. The major requirement for interoperability is that databank designers organize data along common rules: in 2009, the Darwin Core version, which defines minimum standard data (with glossary and synonymy) related with biodiversity, has been internationally adopted for that purpose.
To sum up, accessibility, organization of information, and interoperability contribute to improving the usefulness of biodiversity databanks.
2.2.3 What Are Data in Biodiversity Databanks?
Databanks are not just (organized) collections of data, but also participate in the very definition of what may count as data in the knowledge of biodiversity, a point already made by Leonelli (2013b) about the field of plant science. As emphasized above, different elements are called “data” in biodiversity databanks, like gene sequences, species descriptions, taxonomic revisions, and geographical information. What do all these elements have in common? For what reasons can they be defined as “data” and qualify for being included in databanks? We shall present and discuss the following working hypothesis about data in biodiversity databanks: what they have in common is not that they are all basic or fundamental in the same sense (this would correspond to a definition of “data” as intrinsically basic pieces of information), but that they may play the same epistemic role: they can be relied upon in the further steps of a scientific inquiry. This corresponds to a functional analysis of data. In the remaining of this section, we argue in favor of this hypothesis by comparing species descriptions and gene sequences when they are both categorized as “data”.
Let us first indicate easy-to-notice differences between species descriptions and gene sequences. On the one hand, the respective situations of species descriptions and gene sequences among life sciences are utterly different. Whereas many other pieces of biological knowledge depend on species descriptions, the use of gene sequences within the process of knowledge production depends on other elements of knowledge that may be found in genomics, proteomics, the study of gene regulation, phylogenetic history, etc. However, despite this difference, both species descriptions and gene sequences, once they have been validated as bona fide data, can be considered a sound floor on which one can step in order to go on and explore further research topics. This functional way of understanding data may be contrasted, in both cases, with the view according to which data are defined by their simplicity or easiness of acquisition. Neither gene sequences nor species descriptions are simple or easy to acquire. They are both issued from complex processes. First, a gene sequence is the result of an interpretative judgment with respect to the result of a biochemical experience; the judgement is about which nucleotides appear in the sequence and their order. Second, when a gene sequence is integrated in a databank, for instance in BoLD, where it is associated with a species name (which can be temporary), researchers may also provide the file containing information about the relevant genetic material, the Polymerase Chain Reaction (PCR) primers used to generate the sequenced amplicon, the identifier of the specimen, as well as the collection record, i.e., the location of the original specimen in a collection of natural history. These items may help other scientists check whether the sequence has been associated with the right species name. They illustrate how complex the transformation of a gene sequence into useful data is. On the side of species description, it should be emphasized that associating a specimen with a species name is also the result of a complex process of hypothesis assessment. This association itself possesses a hypothetical status, as it can be changed (via taxonomic revision) when a new set of characters is taken into account or when new specimens are collected. The information-processing facilities that are currently operated within databanks allow databank designers to create links between a specimen and the various taxonomic hypotheses (species names) that have been associated with it over time. In the marine field, this revision process can be followed in the literature and more easily in the WoRMS databank (see below).
The above shows that biodiversity data, the components of biodiversity databanks, are not called “data” because of their simplicity or because they are easily obtained. They are bona fide data thanks to the robust scientific processes on which their production relies. This means that even though some of them require complex material devices for their generation or years of scientific education, these processes are judged reliable enough to be bracketed as the inquiry develops. To put it in another way, the components of biodiversity databanks are so firmly established that, even though they do have a hypothetical character, as any item of knowledge within empirical science, this hypothetical character can be ignored as far as we know. For sure, they do not have the same status as proven mathematical theorems, but they are sufficiently well established to count as firm grounds for knowledge production.
For all the above-presented reasons: international effort of standardization, accessibility, interoperability, robust processes of data production, databanks appear as an efficient way to overcome the discrepancy between the reality of biodiversity knowledge, which is heterogeneous and patchy, and the hope that it may become more and more homogeneous and united. In the next part, we present how existing biodiversity databanks are used by scientists in order to make clear in which ways the organizational logic of databanks relates to the dynamics of knowledge development.
3 Uses of Biodiversity Databanks
This section is devoted to studying the various ways scientists, as distinguished from conservation practitioners and policy makers, use biodiversity databanks in order to develop biodiversity knowledge. We try to disclose the requirements useful databanks have to fulfill and how the data they include are operated in the production of knowledge. In Sect. 3.3.1, we describe, based on examples, what scientists do with the data they retrieve from databanks as well as the quick evolution of this scientific practice. In Sect. 3.3.2, we systematically compare catalogs and databanks in order to explore the specificities of the relationships between a databank and its expected users. At last, we try to show the underlying organizational principles of biodiversity databanks and how they may foster the evolution of scientific knowledge.
3.1 What Do Scientists Do with the Data They Retrieve from Biodiversity Databanks?
As explained above, “data” in biodiversity databanks are already complex units of knowledge that are used to work out other types of scientific results, usually more general and systematic. These data are used to express general hypotheses that cannot be formulated unless two important features of data in biodiversity databanks are realized: they have (i) to cover large geographical regions or large taxonomic groups and (ii) to be valid. Scientists interested in biodiversity study, assessment or conservation may be familiar with a taxonomic group or geographical area and have personal estimates of species abundance in this group or of biodiversity in this area; however, they cannot only rely on their personal experience to put forward general hypotheses and submit them to empirical test. Usually, personal experience, however valuable, is not robust enough to allow for hypothesis testing. By contrast, data in databanks possess the quality that personal connection with biodiversity will always lack: they have been validated by the scientific community, and as such, as explained in Sect. 3.2, they can be relied upon to explore new, more general hypotheses and build up quantitative models.
In order to illustrate, first, how databanks provide researchers with valid pieces of knowledge that they can rely upon and second, the quick transformations of this practice, we shall now present a recent databank that has been created at the Paris Muséum National d’Histoire Naturelle. It is called “BasExp”: Databasis for scientific Expeditions (https://expeditions.mnhn.fr/) and has been designed to gather data related to a 40-year-long programme of marine expeditions initiated by Paris Muséum National d’Histoire Naturelle and the Institut de Recherche pour le Développement, first called “Musorstom”, then “Tropical Deep Sea Benthos”. BasExp collects scientific papers, monographs, and reports issued from this programme. It combines information from these papers, books and reports with data relative to the collected specimens that are preserved at the Paris Muséum. It also collects information about the marine expeditions themselves (not only about the scientific information they have contributed to establish), like who was on board, the main objectives of the expedition, its location, origin of funding, sampling sites, quantity of associated publications, etc. Its main purpose is to allow scientists to overcome two major biases affecting the study of biodiversity, the taxon sampling bias, and the geographic sampling bias, by allowing researchers to know more about the context in which specimens have been sampled. The taxon sampling bias is the tendency to focus on a particular group of organisms and ignore organisms from other groups. The geographical sampling bias is the tendency to go again and again in the same geographical areas to sample specimens instead of exploring other areas. For instance, information about the various researchers on board (specialists of fish, of crustaceans, etc.) may reveal why some taxa were more extensively collected (and on the contrary, the absence of any specialist of a given taxon on board, in a given expedition, may explain why no specimen of this taxon was collected). In a similar way, its being funded by the fish industry might explain the over-representation of fish specimens in another expedition, and so on. There is no doubt that it is important to take these factors into account when looking for biodiversity patterns. As BasExp provides researchers with key elements of the context of sampling, it may be an adequate tool to avoid these common biases.
We now turn to the context in which BasExp has been created. This will shed more light on its purpose and potential benefits for researchers. Most first-generation biodiversity databanks have emerged as answers to inventory requirements. The practice of biodiversity inventory is characterized by its being static in the sense that it is blind, by nature, to changes in biodiversity. Inventories may now be considered of limited interest for biodiversity knowledge because changes in biodiversity are currently a major epistemic challenge, either with respect to conservation or in order to assess the effects of climate change. By contrast with inventory-based databanks, more up-to-date databanks aim at tracking biodiversity change. The best way to do so is by connecting several databanks together in order to be able to follow the evolution of spatiotemporal data in as much details as possible. BasExp allows for such connections.
Among other changes in biodiversity databanks, another one is worth mentioning: they are currently evolving toward less taxa-centered architectures. Many databanks, especially those related with natural history museums, are organised along taxonomic group divisions: one databank for flowering plants, one for crustaceans, etc. However, as emphasized above, there is an increasing need to access a synthetic representation of biodiversity that overcomes the intrinsic limits of taxa-oriented databanks. As biodiversity has to be captured according to many different aspects (geographical, dynamical, taxonomic, genetic, etc.) at once, some databanks offer scientists the means to question their data according to several criteria. BasExp nicely illustrates this possibility. In particular, as there has been a huge effort within BasExp to homogenize geographical data about sampling locations, which were difficult to find out and exploit in the past, it provides researchers with a new and long-awaited type of ready-to-use information. Outside BaxExp, geographical information varies in format and degree of precision from one collection to the next within the Paris Muséum. Gathering all information on a given location and standardizing its format is thus an important advance in itself. Moreover, before the establishment of BasExp, each collection databank had its own data system: they did not use the exact same names for expeditions and did not have the same degree of precision for geographic location. By now, BasExp is the geographical data provider for all the Paris Muséum’s collection databanks. Because it is not taxa-centered, BasExp provides researchers with a synthetic representation of what has been studied and what remains to be investigated about deep-sea fauna.
3.2 Databanks vs. Catalogs
In this section, we put forward a systematic comparison between library or collection catalogs and databanks in order to make clear what the specific features of databanks are from the users’ point of view. We shall show that far from being improved catalogs, allowing for gain in time, databanks are flexible tools that play new roles in the development of biodiversity knowledge.
An important difference between databanks and catalogs is that databanks allow for more than one guiding principle with respect to organisation of information. Let us first explain what we mean by a “guiding principle” with respect to organization of information. In a museum of natural history, catalogs are usually designed according to the way specimens’ identifications are produced. By contrast, an internet-connected databank may be organised according to several guiding principles: specimens’ names, geographical localization, gene sequence, date of discovery, name of discoverer, etc. The organisation of information along multiple dimensions, all of them of scientific interest, is an efficient way to disclose connections that remain inaccessible to catalog users. A nice illustration of this important feature of databanks is that whereas the absence of a species at some place cannot be inferred from consulting a catalog, it may be discovered by cleverly questioning relevant databanks. The main reason for that difference is that catalogs based on specimen identifications cannot but register the presence of specimens without leaving any opportunity to discover information about absence, whereas a databank organised by geographical location may disclose information about absence.
Let us mention another difference between databanks and catalogs, which revolves about their users. Usually, the users of a catalog are determined before its implementation. For instance, the users of the catalog of a museum’s collection are often meant to be people working at the museum, most of them taxonomists: catalogs are most devised for local use. By contrast, internet-based databanks are usually used by different users, even more so when they are inter-connected by means of an Information System, namely, a network of devices for the acquisition, organization, storage, and communication of information that is developed and managed by the host institution. The users’ variety forces databanks’ designers to conceive the organisation of information in such a way as to push the boundaries of local use. The needs and interests of local users of a catalog, e.g., the members of a museum’s scientific community, are more easily identified and narrower than the needs and interests of external users. Taking the latter into account forces databanks’ designers to introduce new possibilities of investigation, e.g., new query types or combinations. This is a very difficult task indeed, as emphasized by Leonelli in the case of biomedical databanks: “[i]ncorporating a large variety of possible viewpoints and prospective queries has been, and continues to be, the most complex and labour-intensive task involved in the development of [the databanks]” (Leonelli 2013a). It amounts to try to guess what the new directions of research may be in order to make the databank usable and useful in the future. This anticipatory task can be said central in the conception and design of biodiversity databanks, whose role in the development of biodiversity knowledge will increase. In Sect. 3.3.3, we further explore the implications of the way information is organised within a databank by showing how their underlying organisational principles may foster the evolution of scientific knowledge.
3.3 Databanks’ Organization and the Dynamics of Biodiversity Knowledge
The principles and functioning of databanks are too often ignored by philosophers of science, who tend to view them as black boxes, whereas looking inside allows one to discover valuable information about the way scientific knowledge is produced on a daily basis. This is why we aim at opening these black boxes and describe their internal functioning in order to show how information is acquired and transformed within them. We will illustrate our claims about the way data are typified and organised by a series of examples.
Each databank’s organization obeys a dominant organisational law that is based on a specific type of information. For instance, as mentioned above, some databanks are centered on taxa whereas others are centered on geographical areas: these are examples of types of information. Other types of information include pictures, taxonomic papers presenting taxonomic descriptions, papers presenting taxonomic revisions, or genetic sequences. It is worth emphasizing that the notion of “type of information” we introduce here is defined with respect to the organisation of biodiversity databanks. For sure, pictures of specimens and taxonomic papers may contain information on the same organisms; however, the information contained in a picture will not play the same role in the process of knowledge-production as the information contained in a taxonomic paper. The databank user will not use a picture in the same way she uses a species description. This is why our notion of type of information is not defined with respect to the item in the world that the information is about, but with respect to the way the information is used by the databank’s user.
Our notion of type of information allows us to establish a classification of biodiversity databanks into taxonomy-oriented, geography-oriented, picture-oriented, revision-oriented, etc. databanks. The type of information that associates a given databank to a class governs what we call the “organisational law” that defines a specific kind of links, within the databank, between the type of information that is the most important within it and other types for information. For instance, the organisational law of a taxonomy-centered databank relates taxonomic data with geographical data, data about endemism, genetic data, etc. The organisational law thus sets apart a center and a periphery within the databank. Center and periphery are defined relative to types of information, as defined above.
Let us make clear how this center-periphery organisation is implemented in various examples.
-
(i)
We begin with WoRMS, the World Register of Marine Species (http://www.marinespecies.org/, already mentioned above). The organisational law for this databank connects original and current species names. As taxonomic revision is at the heart of taxonomic practice, it is highly important to keep track of revisions in order to avoid collective oversight, which would have devastating consequences. WoRMS contains, for each species name, the list of its revisions, dates, and names of associated taxonomists. The use of WoRMS is name-oriented; it delivers information on the sequence of past revisions. It is an international databank. Peripheral information is, e.g., the geographical distribution of the species.
-
(ii)
The OBIS geographical databank (Ocean Biogeographic Information System, http://www.iobis.org/) is sampling-event and geography-oriented, and specific to marine species; it also provides physical, chemical and topographic information on precise locations where specimens have been observed or sampled.
-
(iii)
GBIF (Global Biodiversity Information Facility, http://www.gbif.org/) is a sampling-event databank mapping signalization of species (observation- or sample-event) all around the world.
-
(iv)
By contrast with GBIF, let us mention INPN (Inventaire National du Patrimoine Naturel, https://inpn.mnhn.fr/accueil/index), which also maps signalization of species (observation- or sample-event) on the French territory, with an emphasis on landscape preservation.
-
(v)
BoLD, already mentioned above, combines genetic sequences with information on individual specimens. It is important to emphasize the differences between BoLD and GenBank, another gene-centered databank (see (vi)). Contrary to BoLD, GenBank is centered on the link between genetic sequences and scientific publications. There is no link with actual specimens within GenBank. By contrast, the users of BoLD look for a genetic sequence and the associated species name and may check themselves, by performing standardized experiments, whether the stored genetic sequence (1) has been correctly established and (2) is associated with the right species name. The users of GenBank are not given this possibility because GenBank provides them with a link between a genetic sequence and a scientific paper; as a result, they cannot but trust the authors of the paper with respect to the validity of the gene sequence and associated species name. By contrast, the genetic sequences in BoLD may be not published (Ratnasingham and Hebert 2007). The difference between BoLD and GenBank illustrates an important division among biodiversity databanks: those that provide links to actual specimens, allowing data-checking by the users, and those that provide links to publications or sampling-events, which oblige the users to trust the original information providers.
-
(vi)
The users of GenBank (https://www.ncbi.nlm.nih.gov/genbank/) are looking for comparisons between their own organisms of interest and model organisms whose genomes have been sequenced and included in the databank. It includes all the sequences that have been published in scientific papers.
-
(vii)
Let us end this list with two examples from the Paris Muséum National d’Histoire Naturelle. COLLECTIONS is the databank of the Muséum’s collections, whose functioning is explained here: http://collections.mnhn.fr/wiki/Wiki.jsp?page=Publication_Internet_en. Each museum has several collection databanks for fish, mollusks, crustaceans, etc. They are specimen-oriented: the users look for a specimen’s number (called “voucher ID”) or a name, in order to get information thereon (nomenclatural status: holotype, paratype, etc.; sample site; name).
-
(viii)
BasExp: As mentioned above, this databank is expedition-oriented: the users look for information related with a given expedition. This databank allows for both non-taxa-oriented research and a global appraisal of an expedition in terms of sampling location, specimens, papers, and reports. It is the repository for geographical data related with expeditions.
Whereas each databank has its own organisational logic, many biodiversity databanks function collectively by means of a series of networks like GBIF. Networks of databanks obey their own laws that govern data flow. Usually, data flow runs from the most standardized and best validated data to lesser controlled databanks. Data flow is thus a powerful way of enhancing and homogenizing standards because downstream databanks, when they happen to become upstream relative to other, more local databanks, have to strengthen the reliability and accessibility of their data. Establishing a connection between two biodiversity databanks is indeed a common way to fill the gaps within each. It also contributes to improve data validation and warrant traceability: as emphasized by Costello and Vanden Berghe (2006), “[g]lobal databases that integrate information on species force the development of standard classifications”.
Databank networking has a further important effect at a higher level of knowledge production: it makes clear that some areas of knowledge that had been considered independent beforehand actually entertain epistemic relationships that are now considered as pivotal. For instance, the links between gene sequencing and species identification, implemented in BoLD, has only been brought to light recently (compared with the long history of species identification and the less long, but still not so recent, history of gene sequencing). Realizing that gene sequencing may facilitate species identification has been a side-effect of developing genetic databanks and databanks networks. BoLD’s strength, in this context, is to allow data-checking by providing links with actual specimens (which do not change over time), whereas other databanks depend on taxonomic descriptions, which are hypothetical and susceptible to be transformed.
Another important tendency in the evolution of the way biodiversity databanks get organised is the gradual expansion of the domain of relevant data. Many elements that were considered irrelevant for biodiversity knowledge are now emerging as constitutive, and thus worth collecting, taking care of, and connecting with other types of data. For instance, BasExp includes information on the extraction device of marine specimens, which it relates with sampling sites as well as with digital pictures illustrating the collected specimens in their substrate. The pictures shot during the expeditions have long been difficult to access by scientists who were not on board; now, their status as vehicles of knowledge has completely changed in the last few years as they became easily accessible within databanks and provide irreplaceable information about either the specimen’s environment or the context of its extraction. BasExp thus nicely illustrates a trend that has been identified by Leonelli (2013a), who emphasizes that the databanks she examines (The Arabidopsis Information Resource, TAIR and the cancer Biomedical Informatics Grid, caBIG) progressively include elements that were not considered important, like archives of data provenance (the methods and instruments originally used to generate data) and links to biological materials. We cannot but agree with Leonelli’s statement that “setting up and updating these resources occupies much of curators’ time and creative efforts”.
A last point has to be made about the connection between the development of biodiversity databanks and the dynamics of biodiversity knowledge. It is about the pace of transformation. The different features of biodiversity databanks do not usually evolve at the same rate, which dictates a mandatory upgrading process. For instance, paper catalogs of large natural history museums have usually not been computerized at once, but rather step by step. Now, each step in this process of computerization took place within its own technological and scientific context, involving innovations that the next steps had to catch up with. Evolving biodiversity databanks is thus no linear process. Some events had large interfering consequences, like the establishment of the BoLD consortium (as described here: http://www.barcodeoflife.org/content/about/what-cbol). As soon as Barcoding of Life proved a useful and fruitful endeavour, each biodiversity databank had to take this program into account, which meant huge adaptive changes. Adapting databanks moreover involves satisfying basic requirements of cumulativeness, editability, and interoperability, as these requirements ensure homogeneous development.
In this section, we have presented and discussed the logic governing the organization of biodiversity databanks by means of examples. We have also emphasized the importance of their interconnection into networks and the interplay between their evolution and the changes within biodiversity knowledge. As transformation is pivotal in any reflection on databanks, we now turn briefly to the ways biodiversity databanks may improve in the future.
4 On the Properties of Useful Biodiversity Databanks: Concluding Remarks
Let us recall that, in this paper, we call a biodiversity databank “useful” when it provides scientists, managers of assessment programmes, and conservationists with the means to successfully achieve their aims. In this section, we present two types of requirements a biodiversity databank has to satisfy in order to be useful in this sense. The first type is more on the technical side (although it is not content-independent) whereas the second is linked to the distinguishing features of biodiversity knowledge.
As mentioned above, a few basic requirements have to be met in any databank that is meant to be used by scientists. First, data have to be standardized. Standardization involves defining different data types (i.e., building up a glossary) in order to optimize interoperability among databanks. We can now bring to light the peculiar difficulties raised by this operation by coming back to the example of the integration of non-taxonomic data in a biodiversity databank, namely, data coming from biological and ecological descriptions (cf. Sect. 3.2.2). In order to include biological and ecological traits within a databank for marine species, the main challenge is to identify which traits are (1) useful and (2) available to researchers and conservationists. This is the first step of data standardization, which obliges databank designers to struggle with linguistic subtleties:
For example, “littoral” habitat can be the marine zone between the low and high tide marks, extend to the continental shelf and include coastal river catchments, and refer to the edge of freshwater lakes. The lack of standard use of terms can compromise the bringing together of this knowledge from different sources. (Costello et al. 2015).
We see in this example that standardization forces databank designers to formulate precise definitions of the terms they choose to use within the databank, as well as to determine measurement units. This is a crucial, non-trivial step in the elaboration of the databank. It is based on the previous identification of the set of terms that are used in other databanks and in the relevant literature. Standardization is not always possible, however: when the way data are produced remains unknown, i.e., when the databank designer does not know whether they are primarily found in scientific papers, reports, other databanks, or unpublished sources, they cannot possibly be standardized because standardization involves checking the validity of data. However, when data are from unknown origin, their validity cannot be properly checked. That is the reason why working groups for data standardization must involve scientists: standardization is knowledge production.
The most important requirement to build up a useful databank is that data have to be validated: they must be submitted to a process that provides scientists with the same kind of warrant as the peer review process so that they can trust the elements they retrieve from databanks. When checking the validity of data, users are assisted by the meta-data that provide them with contextual information allowing them to both evaluate reliability and quality of data. Meta-data are sets of data that accompany the constitutive data of a databank: for instance, in a taxonomy-oriented databank, the constitutive data may be species descriptions and associated meta-data geographical information, gene sequences, pictures, location of holo- and para-types, etc. When links to actual specimens are provided, databank users can moreover check themselves the validity of the data they are interested in. These links are thus an important way to enhance the collective process of data validation, and, as a result, the overall scientific quality of biodiversity data and databanks.
The usefulness of a databank is conditioned by the basic requirements so far illustrated and by the definition of possible queries. By listing the words or expressions that constitute well-defined queries, databank designers identify and delineate the possible uses of the tool. As mentioned above, the databank designers’ role is to guess the future uses of the databank and to anticipate the developmental paths of biodiversity knowledge. This can only be done by people in close contact with on-going research, assessment, and conservation practices: even though it may not always result in the publication of scientific papers, databank design and maintenance is genuine scientific work because it implies being well aware of the scientific state of the art at a given time, of how the relevant scientific community assesses the various scientific hypotheses at stake, and of the emerging links among different fields of research.
Even though biodiversity databanks already have a history, they are recent tools compared to the old practice of publication of papers and monographs. A major challenge for biodiversity databanks is to become a part in a set of older practices that themselves transform at a quick pace. This means that biodiversity databanks should connect their contents with other, older epistemic practices. More precisely, they should complement, rather than replace, more traditional reservoirs of biodiversity knowledge, like collections of natural history, because reliable biodiversity knowledge requires links with actual specimens. This is an important and difficult task for databanks designers and associated computer scientists and engineers.
Notes
- 1.
In order to provide the reader with evidence for this claim, we searched for “earth worm” on current search engines and found out that, outside taxonomy, scientific publications about earth worms exceptionally mention James et al. 2010 paper and the revision it contains. This suggests that non-taxonomists simply do not pay attention to the way scientific knowledge develops and changes within taxonomy and that taxonomic revisions are not generally considered mandatory elements within their own knowledge of biodiversity, whereas they play a central role within taxonomic knowledge of biodiversity.
- 2.
The relationship between the most developed attempt to provide ecology with a theory, namely the Neutral Theory of Ecology (Hubbell 2001), illustrates this point: the Neutral Theory is not yet unified with evolutionary theory, according to its proponent himself.
- 3.
The fact that knowledge of taxonomic groups is not uniformly spread is a major problem for biodiversity assessment and conservation (see e.g. http://www.iucnredlist.org/about/summary-statistics).
- 4.
https://www.cbd.int/convention/text/ (On the CBD, see also Oksanen and Vuorisalo in this volume).
References
Baetu, T. M. (2012). Genes after the human genome project. Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences, 43, 191–201.
Bastow, R., & Leonelli, S. (2010). Sustainable digital infrastructure. EMBO Reports, 11(10), 730–735.
Bouchet, P. (2006). The magnitude of marine biodiversity. In C. M. Duarte (Ed.), The exploration of marine biodiversity. Scientific and technological challenges (pp. 31–62). Bilbao: Fundacion BBVA.
Carlson, E. A. (1991). Defining the gene: An evolving concept. The American Journal of Human Genetics, 49, 475–487.
Costello, M. J., & Vanden Berghe, E. (2006). Ocean biodiversity informatics: A new era in marine biology research and management. Marine Ecology Progress Series, 316, 203–214.
Costello, M. J., Claus, S., Dekeyzer, S., Vandepitte, L., Tuama, É. Ó., Lear, D., & Tyler-Walters, H. (18 août 2015). Biological and ecological traits of marine species. PeerJ, 3, e1201. https://doi.org/10.7717/peerj.1201.
Covitz, P. A. (2004). Cruising the cancer biomedical informatics grid caBIG: From village to city (caBIG Workspace and Working Group Kickoff meeting, 2004).
Falk, R. (1986). What is a gene? Studies in the History and Philsophy of Science, 17, 133–173.
Fogle, T. (2000). The dissolution of protein coding genes in molecular biology. In P. Beurton, R. Falk, & H.-J. Rheinberger (Eds.), The concept of the gene in development and evolution. Historical and epistemological perspectives (pp. 3–25). Cambridge: Cambridge University Press.
Gerstein, M. B., Bruce, C., Rozowsky, J. S., Zheng, D., Du, J., Korbel, J. O., Emanuelsson, O., Zhang, Z. D., Weissman, S., & Snyder, M. (2007). What is a gene, post-ENCODE? History and updated definition. Genome Research, 17, 669–681.
Grehan, J. R. (1993). Conservation biogeography and the biodiversity crisis: A global problem in space/time. Biodiversity Letters, 1(5), 134–140.
Hubbell, S. P. (2001). The unified neutral theory of biodiversity and biogeography. Princeton: Princeton University Press.
James, S. W., Porco, D., Decaëns, T., Richard, B., & Rougerie, R. (2010). DNA barcoding reveals cryptic diversity in Lumbricus terrestris L., 1758 (Clitellata): Resurrection of L. herculeus (Savigny, 1826). PLoS One, 5(12), e15629. https://doi.org/10.1371/journal.pone.0015629.
Kitcher, P. (1982). Genes. British Journal for the Philosophy of Science, 33, 337–359.
Leonelli, S. (2009). The impure nature of biological knowledge. In H. de Regt, S. Leonelli, & K. Eigner (Eds.), Scientific understanding: Philosophical perspectives. Pittsburgh: Pittsburgh University Press.
Leonelli, S. (2010). The commodification of knowledge exchange: Governing the circulation of biological data. In H. Radder (Ed.), The commodification of academic research. Pittsburgh: University of Pittsburgh Press.
Leonelli, S. (2013a). Global data for local science: Assessing the scale of data infrastructures in biological and biomedical research. BioSociety, 8(4), 449–465.
Leonelli, S. (2013b). Integrating data to acquire new knowledge: Three modes of integration in plant science. Studies in History and Philosophy of Biological and Biomedical Sciences, 44(4 Pt A), 503–514. https://doi.org/10.1016/j.shpsc.2013.03.020.
Leonelli, S. (2016). Data-centric biology. A philosophical study. Chicago: The University of Chicago Press.
OECD. (1999). OECD megascience working group – Biological informatics – Final report. 74 pp. Organisation for Economic Co-operation and Development. Available online at http://www.oecd.org/dataoecd/24/32/2105199.pdf
Olson, D. M., Dinerstein, E., Powell, G. V. N., & Wikramanayake, E. D. (2002). Conservation biology for the biodiversity crisis. Conservation Biology, 16(1), 1–3.
Ratnasingham, S., & Hebert, P. D. N. (2007). BOLD: The barcode of life data system (http://www.barcodinglife.org). Molecular Ecology Notes, 7(3), 355–364.
Richard, B., Decaëns, T., Rougerie, R., James, S. W., & Porco, D. (2009). Re-integrating earthworm juveniles into soil biodiversity studies: Species identification through DNA barcoding. Molecular Ecology Ressources, 10, 606–614.
Sarkar, S. (2016). Ecology. The stanford encyclopedia of philosophy. In E. N. Zalta (Ed.). https://plato.stanford.edu/archives/win2016/entries/ecology/
Singh, J. S. (2002). The biodiversity crisis: A multifaceted review. Current Science, 82(6), 638–647.
Sterelny, K., & Kitcher, P. (1988). The return of the gene. Journal of Philosophy, 85, 339–360.
Takacs, D. (1996). The idea of biodiversity: Philosophies of paradise. Baltimore: Johns Hopkins University Press.
Waters, K. C. (1994). Genes made molecular. Philosophy of Science, 61, 163–185.
Western, D. (1992). The biodiversity crisis: A challenge for biology. Oikos, 63(1), 29–38.
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Döring, M., Giovanni, R., Robertson, T., & Vieglais, D. (2012). Darwin core: An evolving community-developed biodiversity data standard. PLoS One, 7(1), e29715.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2019 The Author(s)
About this chapter
Cite this chapter
Barberousse, A., Bary, S. (2019). Marine Biodiversity Databanks. In: Casetta, E., Marques da Silva, J., Vecchi, D. (eds) From Assessing to Conserving Biodiversity. History, Philosophy and Theory of the Life Sciences, vol 24. Springer, Cham. https://doi.org/10.1007/978-3-030-10991-2_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-10991-2_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-10990-5
Online ISBN: 978-3-030-10991-2
eBook Packages: Religion and PhilosophyPhilosophy and Religion (R0)