Abstract
Lung cancer is a leading cause of mortality and morbidity for patients suffering from Chronic Obstructive Pulmonary Disease (COPD). Both the presence of visually assessed emphysema on CT scans and abnormal pulmonary function tests are associated with the development of lung cancer. Based on recent results showing that convolutional neural networks (CNNs) applied to CT scans can predict spirometrically-defined COPD (\(\frac{FEV_{1}}{FVC}<0.7\)), we hypothesized that CNN-based classification of COPD and emphysema is predictive of lung cancer development in the National Lung Cancer Screening (NLST) cohort. We trained spirometric COPD and visual emphysema CNN classifiers using data from the COPDGene study. The classifiers were then used to generate COPD and emphysema scores (\(CS_{CNN}\) and \(ES_{CNN}\), respectively) on 7347 CT scans from the NLST study. Cox proportional hazards regression was used to model the effects of \(CS_{CNN}\), \(ES_{CNN}\), age, body mass index, education, gender, smoking pack-years, and years since smoking cessation on lung cancer diagnosis. It was found that, individually, both \(CS_{CNN}\) and \(ES_{CNN}\) were statistically significant predictors (p < 0.000 and p < 0.000, respectively) of lung cancer diagnosis hazard.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Chronic Obstructive Lung Disease (COPD), an inflammatory lung disease resulting in pulmonary airflow obstruction, is projected to be the fourth leading cause of death in the world by 2030 [1]. COPD is typically diagnosed using spirometry (i.e. pulmonary function tests, PFTs), with a forced expiratory volume in 1 second to forced vital capacity ratio (\(\frac{FEV_{1}}{FVC}\)) less than 70% being considered a COPD diagnosis [1]. Emphysema, a sub-type of COPD that involves the thinning and destruction of the alveoli, is one of the diseases comprising COPD. Emphysema presence and severity is typically assessed by visual reading of thoracic computed tomography (CT) scans.
Lung cancer has been shown to be associated with spirometrically defined COPD (sCOPD), with Young et al. reporting a two-fold increase in lung cancer incidence among patients in the National Lung Cancer Screening Trial (NLST) who had COPD [2]. The presence of visually assessed emphysema on CT scans is also associated with lung cancer diagnosis, as a meta-analysis conducted by Smith et al. showed that the presence vs. absence of visual emphysema on CT resulted in a lung cancer diagnosis odds ratio of 3.50 [3].
It is therefore possible that using PFT and/or CT imaging data may allow for more accurate lung cancer risk stratification, which could improve lung cancer screening inclusion criteria and/or be used to help motivate patients to quit smoking [5]. Unfortunately, PFTs and visual assessment of emphysema are not always available due to the associated costs. In addition, visual assessment of emphysema is subjective and thus suffers from high intra and inter-reader variability [6].
It was recently shown that Convolutional Neural Networks (CNNs) can be used to train end-to-end CT-based classifiers of clinical COPD outcomes [7] such as GOLD stage, exacerbation frequency, and mortality. Based on these results, we hypothesized these techniques could used to improve lung cancer risk modeling without the need for PFTs or visual assessment of emphysema.
In this work, we present a CT-based CNN classification workflow for assessment of sCOPD and visual emphysema and show that classification results produced by the CNNs are predictive of lung cancer diagnosis hazard in the NLST cohort.
2 Methods
2.1 Data
The CNNs were trained using image and clinical data from the Genetic Epidemiology of COPD (COPDGene) study [8]. CT scans from the baseline image collection and 5-year follow-up were available for training and validation. Only scans reconstructed using a smooth kernel (GE Standard, Siemens B31f, or Philips B) were used in this study.
The CNN models trained on COPDGene data were applied to CT scans from the NLST. We processed low-dose CT scans from the NLST that were reconstructed with a Siemens B30f, GE Standard, Philips B, or Toshiba FC10 kernel, and that had a slice thickess of 2.5 mm or less. After accounting for missing clinical data and failed image processing, this resulted in 7347 datasets. 2694 of these datasets had associated spirometry data.
2.2 CNN Architecture
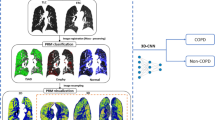
Image Processing. High resolution CT volumes are too large to train and process on current graphics processing units (GPUs). Similar to [7], we developed a data reduction strategy that used a subset of image slices for training and processing. A set of 8 axial slices, each down-sampled from 512\(\,\times \,\)512 to 256\(\,\times \,\)256 pixels, were randomly sampled from equally sized “zones” of the lung and combined into a single image montage (Fig. 1).
CNN Configuration. The CNN configuration is presented in Table 1. Both sCOPD and visual emphysema classifiers used this configuration for training and testing. The CNN was implemented in PyTorch and trained using stochastic gradient descent with a cross-entropy loss function, Nesterov momentum of 0.9, a learning rate of 0.001, and a batch size of 32.
Training. For the sCOPD classifier, subjects were classified as having COPD if they were in Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage 1 or greater. GOLD stage 0 and PRISM subjects were classified as not having COPD. 3750 subjects were used for training. Due to the to use of random slices for processing each CT scan, data augmentation was used to increase the size of the training dataset from 3750 to 15000 by generating four different slice configurations for each subject. 5-year follow-up CT scans were used for training validation.
The emphysema classifier was trained using visual centrilobular emphysema classifications. A subset of the baseline COPDGene CT scans were visually scored by two analysts using the Fleischner Society classification system. Emphysema was classified as centrilobular (trace, mild, moderate, confluent, and advanced destructive emphysema), panlobular, and paraseptal (mild or substantial). A two-category classifier was generated that classified an image as having visual emphysema if it contained mild, moderate, confluent, or advanced destructive centrilobular emphysema, and no emphysema otherwise. Visual reads that were not agreed upon by both analysts were not used for training. A total of 875 subjects were used for training, resulting in 3500 training images after using the data augmentation strategy outlined above. 3500 datasets from different subjects were used for training validation.
CNN Validation and Testing. Following training, the CNN models were used to create sCOPD and emphysema classification probabilities (i.e. classification scores  ,
,  , respectively) by taking the exponential of the model output. Classification scores were assigned to a binary category by thresholding at 0.5 (e.g. \(CS_{CNN}>0.5 \rightarrow \) sCOPD, \(CS_{CNN}\le 0.5 \rightarrow \) NO sCOPD). Validation and test accuracy was computed as the percentage of correct classifications.
, respectively) by taking the exponential of the model output. Classification scores were assigned to a binary category by thresholding at 0.5 (e.g. \(CS_{CNN}>0.5 \rightarrow \) sCOPD, \(CS_{CNN}\le 0.5 \rightarrow \) NO sCOPD). Validation and test accuracy was computed as the percentage of correct classifications.
The sCOPD classifier was further validated in a subset of NLST images (2694 subjects) with spirometry data available. Radiologist generated visual emphysema classification, however, was not available for the NLST datasets.
2.3 Statistical Analysis
Cox proportional hazard models were used to test the association between \(CS_{CNN}\) and \(ES_{CNN}\) and time-to-event of lung cancer diagnosis incidence. Participants were censored at 6 years of follow-up. Regression models were adjusted for covariates known to be associated with cancer development including age, gender, body mass index, pack-years, and time since smoking cessation. Three models were generated: One with both \(CS_{CNN}\) and \(ES_{CNN}\) scores included as continuous variables, one with only the \(CS_{CNN}\) score included as a continuous variable, and one with only the \(ES_{CNN}\) score included as a continuous variable.
Kaplan-Meier curves were also generated for \(CS_{CNN}\) and \(ES_{CNN}\) classifiers for subjects that fell above and below the median classification scores within the NLST cohort (Fig. 2).
Both Cox proportional hazards regression and Kaplan-Meier analysis were implemented in Python using the lifelines package.
3 Results
3.1 Validation and Test Accuracy
Validation accuracy for the COPDGene cohort and test accuracy for the NLST cohort are shown in Table 2. The validation accuracy of the \(CS_{CNN}\) classifier was almost exactly the same as reported for test subset in [7]. It should also be noted that there was almost no decrease in the sCOPD classification accuracy when going from the COPDGene to the NLST scans, despite that fact that the patient cohorts and CT image acquisition and reconstruction parameters were different.
3.2 Statistical Analysis
Results for each Cox model are shown in Table 3. When \(CS_{CNN}\) and \(ES_{CNN}\) were not combined into a single model, both were statistically significant (p < 0.000) predictors of lung cancer diagnosis hazard. When combined in a single model, however, the statistical significance of the \(CS_{CNN}\) and \(ES_{CNN}\) classification scores decreased (to \(p=0.0195\) and \(p=0.0598\), respectively).
4 Discussion and Conclusion
The relationship between objective quantitative CT-based assessment of emphysema (i.e. percent low-attenuation area, %LAA) and lung cancer has been shown to be either weak [9] or non-existent [3, 10, 11], despite an association between visual emphysema and lung cancer. In this work, we showed that CNN-based probability scores of spirometrically defined COPD and visual emphysema were both statistically significant predictors of lung cancer diagnosis hazard in the NLST cohort. An encouraging result of this work is that, although the sCOPD classifier was trained on full-dose CTs from the COPDGene study, the validation accuracy of the classifier decreased only 1.5% when applied to low-dose CTs from the NLST, which is evidence that the classifier was robust and not overfit to the training data.
The CNN architecture presented in this work uses only a subset of axial slices from a high-resolution CT image. A potential benefit of this architecture is that it might be possible to obtain accurate classification of sCOPD and/or emphysema from “incremental” CT scans (scans acquired with large spacing between slices), which when used in combination with low-tube currents, would allow for screening of COPD at very low x-ray doses. Another benefit of the image processing workflow is that, due to the use of random slices within lung zones, it may be possible to increase the accuracy of the classifier estimates by ensembling the results from multiple configurations.
A limitation of this preliminary work is that CNN models were only trained to classify COPD and emphysema as binary categories even though more granular data was available (e.g. GOLD 0–4 and Fleischner society emphysema classifications). Additionally, the variation in CNN scores obtained using different random slice configurations from the same image should be characterized to help understand the classification repeatability. Finally, a comparison with the performance of other quantitative CT-based COPD metrics (e.g. LAA-950 or Perc15) is of particular interest. Future work will focus on addressing these issues.
In conclusion, we trained CNNs to classify COPD and emphysema presence from CT images, and showed that the classification probabilities were statistically significant predictors of lung cancer diagnosis hazard.
References
Cruz, A.A.: Global surveillance, prevention and control of chronic respiratory diseases: a comprehensive approach. World Health Organization (2007)
Young, R.P., et al.: Airflow limitation and histology shift in the national lung screening trial. The NLST-ACRIN cohort substudy. Am. J. Respir. Criti. Care Med. 192(9), 1060–1067 (2015)
Smith, B.M., Pinto, L., Ezer, N., Sverzellati, N., Muro, S., Schwartzman, K.: Emphysema detected on computed tomography and risk of lung cancer: a systematic review and meta-analysis. Lung Cancer 77(1), 58–63 (2012)
National Lung Screening Trial Research Team: Reduced lung-cancer mortality with low-dose computed tomographic screening. New Engl. J. Med. 365(5), 395–409 (2011)
McClure, J.B.: Are biomarkers a useful aid in smoking cessation? A review and analysis of the literature. Behav. Med. 27(1), 37–47 (2001)
Bankier, A.A., De Maertelaer, V., Keyzer, C., Gevenois, P.A.: Pulmonary emphysema: subjective visual grading versus objective quantification with macroscopic morphometry and thin-section CT densitometry. Radiology 211(3), 851–858 (1999)
González, G., et al.: Disease staging and prognosis in smokers using deep learning in chest computed tomography. Am. J. Respir. Crit. Care Med. 197(2), 193–203 (2018)
Regan, E.A., et al.: Genetic epidemiology of COPD (COPDGene) study design. COPD J. Chronic Obstr. Pulm. Dis. 7(1), 32–43 (2011)
Gierada, D.S., et al.: Quantitative CT assessment of emphysema and airways in relation to lung cancer risk. Radiology 261(3), 950–959 (2011)
Wilson, D.O., et al.: Association of radiographic emphysema and airflow obstruction with lung cancer. Am. J. Respir. Crit. Care Med. 178(7), 738–744 (2008)
Maldonado, F., Bartholmai, B.J., Swensen, S.J., Midthun, D.E., Decker, P.A., Jett, J.R.: Are airflow obstruction and radiographic evidence of emphysema risk factors for lung cancer?: a nested case-control study using quantitative emphysema analysis. Chest 138(6), 1295–1302 (2010)
Acknowledgements
The authors thank the National Cancer Institute for access to NCI’s data collected by the National Lung Screening Trial. The statements contained herein are solely those of the authors and do not represent or imply concurrence or endorsement by NCI. This work was supported by NIH grant 2R44CA203050-02. The COPDGene study is supported by NIH Grant Numbers R01 HL089897 and R01 HL089856, and is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, Sunovion and GlaxoSmithKline.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Hatt, C., Galban, C., Labaki, W., Kazerooni, E., Lynch, D., Han, M. (2018). Convolutional Neural Network Based COPD and Emphysema Classifications Are Predictive of Lung Cancer Diagnosis. In: Stoyanov, D., et al. Image Analysis for Moving Organ, Breast, and Thoracic Images. RAMBO BIA TIA 2018 2018 2018. Lecture Notes in Computer Science(), vol 11040. Springer, Cham. https://doi.org/10.1007/978-3-030-00946-5_30
Download citation
DOI: https://doi.org/10.1007/978-3-030-00946-5_30
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00945-8
Online ISBN: 978-3-030-00946-5
eBook Packages: Computer ScienceComputer Science (R0)