Abstract
Multi-modal neuroimages (e.g., MRI and PET) have been widely used for diagnosis of brain diseases such as Alzheimer’s disease (AD) by providing complementary information. However, in practice, it is unavoidable to have missing data, i.e., missing PET data for many subjects in the ADNI dataset. A straightforward strategy to tackle this challenge is to simply discard subjects with missing PET, but this will significantly reduce the number of training subjects for learning reliable diagnostic models. On the other hand, since different modalities (i.e., MRI and PET) were acquired from the same subject, there often exist underlying relevance between different modalities. Accordingly, we propose a two-stage deep learning framework for AD diagnosis using both MRI and PET data. Specifically, in the first stage, we impute missing PET data based on their corresponding MRI data by using 3D Cycle-consistent Generative Adversarial Networks (3D-cGAN) to capture their underlying relationship. In the second stage, with the complete MRI and PET (i.e., after imputation for the case of missing PET), we develop a deep multi-instance neural network for AD diagnosis and also mild cognitive impairment (MCI) conversion prediction. Experimental results on subjects from ADNI demonstrate that our synthesized PET images with 3D-cGAN are reasonable, and also our two-stage deep learning method outperforms the state-of-the-art methods in AD diagnosis.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Multi-instance Neural Networks
- Mild Cognitive Impairment (MCI)
- Underlying Relevance
- Image Synthesis Model
- Conv Layers
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Structural magnetic resonance imaging (MRI) and positron emission tomography (PET) have been widely used for diagnosis of Alzheimer’s disease (AD) as well as prediction of mild cognitive impairment (MCI) conversion to AD. Recent studies have shown that MRI and PET contain complementary information for improving the performance of AD diagnosis [1, 2].
A common challenge in multi-modal studies is the missing data problem [3, 4]. For example, in clinical practice, subjects who are willing to be scanned by MRI may reject PET scans, due to high cost of PET scanning or other issues such as concern of radioactive exposure. In the baseline Alzheimer’s Disease Neuroimaging Initiative (ADNI-1) database, only approximately half of subjects have PET scans, although all 821 subjects have MRI data. Previous studies usually tackle this challenge by simply discarding subjects without PET data [5]. However, such simple strategy will significantly reduce the number of training subjects for learning the reliable models, thus degrading the diagnosis performance.
To fully utilize all available data, a more reasonable strategy is to impute the missing PET data, rather than simply discarding subjects with missing PET data. Although many data imputing methods have been proposed in the literature [3], most of them focus on imputing missing feature values that are defined by experts for representing PET. Note that, if these hand-crafted features are not discriminative for AD diagnosis (i.e., identifying AD patients from healthy controls (HCs)), the effect of imputing these missing features will be very limited in promoting the learning performance. Therefore, in this work, we focus on imputing missing PET images, rather than hand-crafted PET features.
Recently, the cycle-consistent generative adversarial network (cGAN) [6] has been successfully applied to learning the bi-directional mappings between relevant image domains. Since MR and PET images scanned from the same subjects have underlying relevance, we resort to cGAN to learn bi-directional mappings between MRI and PET, through which missing PET scan can be then synthesized based on its corresponding MRI scan.
Specifically, we propose a two-stage deep learning framework to employ all available MRI and PET for AD diagnosis, with the schematic illustration shown in Fig. 1. In the first stage, we impute the missing PET images by learning bi-directional mappings between MRI and PET via 3D-cGAN. In the second stage, based on the complete MRI and PET (i.e., after imputation), we develop a landmark-based multi-modal multi-instance learning method (\(\text {LM}^3\text {IL}\)) for AD diagnosis, by learning MRI and PET features automatically in a data-driven manner. To the best of our knowledge, this is one of the first attempt to impute 3D PET images using deep learning with cycle-consistent loss in the domain of computer-aided brain disease diagnosis.
2 Method
Problem Formulation: Assume \(\left\{ \mathbf {X}_M^i, \mathbf {X}_P^i\right\} _{i=1}^N\) is a set consisting of N subjects, where \(\mathbf {X}_M^i\in \mathcal {X_M}\) and \(\mathbf {X}_P^i\in \mathcal {X_P}\) are, respectively, the MRI and PET data for the \(i^{\text {th}}\) subject. A multi-modal diagnosis model can then be formulated as \(\mathbf {\hat{y}}^i=F\left( \mathbf {X}_M^i,\mathbf {X}_P^i\right) \), where \(\mathbf {\hat{y}}^i\) is the predicted label (e.g., AD/HC) for the \(i^{\text {th}}\) subject. However, when the \(i^{\text {th}}\) subject does not have PET data (i.e., \(\mathbf {X}_P^i\) is missing), the model \(F\left( \mathbf {X}_M^i,-\right) \) cannot be executed. An intuitive way to address this issue is to use data imputation, e.g., to predict a virtual \(\mathbf {\hat{X}}_P^i\) using \(\mathbf {X}_M^i\), considering their underlying relevance. Letting \(\mathbf {\hat{X}}_P^i= G\left( \mathbf {X}_M^i\right) \) denoting data imputation with the mapping function G, the diagnosis model can then be formulated as
Therefore, there are two sequential tasks in the multi-modal diagnosis framework based on incomplete data, i.e., (1) learning a reliable mapping function G for missing data imputation, and (2) learning a classification model F to effectively combine complementary information from multi-modal data for AD diagnosis. To deal with the above two tasks sequentially, we propose a two-stage deep learning framework, consisting of two cascaded networks (i.e., 3D-cGAN and LM\(^3\)IL as shown in Fig. 1), with the details given below.
Stage 1: 3D Cycle-consistence Generative Adversarial Network (3D-cGAN). The first stage aims to synthesize missing PET by learning a mapping function \(G{:}~\mathcal {X_M}\rightarrow \mathcal {X_P}\). We require G to be a one-to-one mapping, i.e., there should exist a reversed function \( G^{-1}{:}~\mathcal {X_P}\rightarrow \mathcal {X_M} \) to keep the mapping consistent.
To this end, we propose a 3D-cGAN model, which is an extension of the existing 2D-cGAN [6]. The architecture of our 3D-cGAN model is illustrated in Fig. 2, which includes two generators, i.e., \(G_1{:}~\mathcal {X_M}\,\rightarrow \,\mathcal {X_P}\) and \(G_2{:}~\mathcal {X_P}\,\rightarrow \,\mathcal {X_M}\) (\(G_2=G_1^{-1}\)), and also two adversarial discriminators, i.e., \( D_{1} \) and \( D_{2} \). Specifically, each generator (e.g., \(G_1\)) consists of three sequential (i.e., encoding, transferring and decoding) parts. The encoding part is constructed by three convolutional (Conv) layers (with 8, 16, and 32 channels, respectively) for extracting the knowledge of images in the original domain (e.g., \(\mathcal {X_M}\)). The transferring part contains 6 residual network blocks [7] for transferring the knowledge from the original domain (e.g., \(\mathcal {X_M}\)) to the target domain (e.g., \(\mathcal {X_P}\)). Finally, the decoding part contains 2 deconvolutional (Deconv) layers (with 8 and 16 channels, respectively) and 1 Conv layer (with one channel) for constructing the images in the target domain (e.g., \(\mathcal {X_P}\)). Besides, each discriminator (e.g., \(D_2\)) contains 5 Conv layers, with 16, 32, 64, 128, and 1 channel(s), respectively. It inputs a pair of real image (e.g., \(\mathbf {X}_P^i\)) and synthetic image (e.g., \(G_1(\mathbf {X}_M^i)\)), and then outputs a binary indicator to tell us whether the real and its corresponding synthetic images are distinguishable (output = 0) or not (output = 1). To train our 3D-cGAN model with respect to \(G_1\), \(G_2\), \(D_1\), and \(D_2\), a hybrid loss function is defined as:
where
are the adversarial loss and cycle consistency loss [6], respectively. The former ensures the synthetic PET images be similar to the real images, while the latter keeps each synthetic PET be consistent with the corresponding real MRI. Parameter \( \lambda \) controls the importance of the consistency.
In our experiments, we empirically set \( \lambda = 10 \), and then trained \(D_1\), \(D_2\), \(G_1\), and \(G_2\) alternatively by minimizing \(-\mathfrak {L}_{g} (G_2, D_1) \), \(-\mathfrak {L}_{g} (G_1,D_2) \), \(\mathfrak {L}_{g} (G_1,D_2)+\lambda \mathfrak {L}_{c} (G_1,G_2) \) and \(\mathfrak {L}_{g} (G_2,D_1)+\lambda \mathfrak {L}_{c} (G_1,G_2) \), iteratively. The Adam solver [8] was used with a batch size of 1. The learning rate for the first 100 epochs was kept as \( 2 \times 10^{-3} \), and was then linearly decayed to 0 during the next 100 epochs.
Stage 2: Landmark-based Multi-modal Multi-Instance Learning (LM\( ^{\mathbf {3}}\)IL) Network. In the second stage, we propose the LM\(^3\)IL model to learn and fuse discriminative features from both MRI and PET for AD diagnosis. Specifically, we extract L patches (with size of \( 24 \times 24 \times 24 \)) centered at L pre-defined disease-related landmarks [9] from each modality. Therefore, for the \(i^{\text {th}}\) subject, we have 2L patches denoted as \(\left\{ {\mathbf {P}_l^i}\right\} _{l=1}^{2L}\), in which the first L patches are extracted from \(\mathbf {X}_M^i\), while the next L patches are extracted from \(\mathbf {X}_P^i\) or \(G_1(\mathbf {X}_M^i)\) when \(\mathbf {X}_P^i\) is missing.
By using \(\left\{ {\mathbf {P}_l^i}\right\} _{l=1}^{2L}\) as the inputs, the architecture of our LM\(^3\)IL model is illustrated in Fig. 3, which consists of 2L patch-level feature extractors (i.e., \( \left\{ {f_l} \right\} _{l=1}^{2L} \)) and a subject-level classifier (i.e., \(f_0\)). All \( \left\{ {f_l} \right\} _{l=1}^{2L} \) have the same structure but different parameters. Specifically, each of them consists of 6 Conv layers and 2 fully-connected (FC) layers, with the rectified linear unit (ReLU) used as the activation function. The outputs of the \(2^{\text {nd}} \), \(4^{\text {th}}\) and \(6^{\text {th}}\) layers are down-sampled by the max-pooling operations. The size of the Conv kernels is \( 3\times 3\times 3 \) in the first two Conv layers, and \( 2\times 2\times 2 \) in the remaining four Conv layers. The number of channels is 32 for the \(1^{\text {st}}\), \(2^{\text {nd}}\) and \(8^{\text {th}}\) Conv layers, 64 for \(3^{\text {rd}}\) and \(4^{\text {th}}\) Conv layers, and 128 for the \(5^{\text {th}}\), \(6^{\text {th}}\) and \(7^{\text {th}}\) layers. Each patch \(\mathbf {P}_l^i\) (\(l\in \{1,\ldots ,2L\}\)) is first processed by the corresponding sub-network \(f_l\) to produce a patch-level feature vector \(\mathbf {O}_l\) (i.e., the outputs of the last FC layer) with 32 elements. After that, feature vectors from all landmark locations in both MRI and PET are concatenated, which are then fed into the subsequent subject-level classifier \(f_0\). The subject-level classifier \(f_0\) consists of 3 FC layers and a soft-max layer, where the first two layers (with the size of 64L and 8L, respectively) aim to learn a subject-level feature representation to effectively integrate complementary information from different patch locations and also different modalities, based on which the last FC layer (followed by the soft-max operation) outputs the diagnosis label (e.g., AD/HC). For the \(i^{\text {th}}\) subject, the whole diagnosis procedure in our LM\(^3\)IL method can be summarized as:
In our experiments, the proposed LM\(^3\)IL model was trained with log loss using the stochastic gradient descent (SGD) algorithm [10], with a momentum coefficient of 0.9 and a learning rate of \(10^{-2}\).
3 Experiments
Materials and Image Pre-processing. We evaluate the proposed method on two subsets of ADNI database [11], including ADNI-1 and ADNI-2. Subjects were divided into four categories: (1) AD, (2) HC, (3) progressive MCI (pMCI) that would progress to MCI within 36 months after baseline time, and (4) static MCI (sMCI) that would not progress to MCI. There are 821 subjects in ADNI-1, including 199 AD, 229 HC, 167 pMCI and 226 sMCI subjects. Also, ADNI-2 contains 636 subjects, including 159 AD, 200 HC, 38 pMCI and 239 sMCI subjects. While all subjects in ADNI-1 and ADNI-2 have baseline MRI data, only 395 subjects in ADNI-1 and 254 subjects in ADNI-2 have PET images.
All MR images were pre-processed via four steps: (1) anterior commissure (AC)-posterior commissure (PC) alignment, (2) skull stripping, (3) intensity correction, (4) cerebellum removal, and (5) linear alignment to a template MRI. Each PET image was also aligned to its corresponding MRI via linear registration. Hence, there is spatial correspondence between MRI and PET for each subject.
Experimental Settings. We performed two groups of experiments in this work. In the first group, we aim to evaluate the quality of the synthetic images generated by 3D-cGAN. Specifically, we train the 3D-cGAN model using subjects with complete MRI and PET scans in ADNI-1, and test this image synthesis model on the complete subjects (with both MRI and PET) in ADNI-2. The averaged peak signal-to-noise ratio (PSNR) is used to measure the image quality of those synthetic PET and MR images generated by our method.
In the second group, we evaluate the proposed \(\text {LM}^3\text {IL}\) method on both tasks of AD classification (AD vs. HC) and MCI conversion prediction (pMCI vs. sMCI) using both real multi-modal images and our synthetic PET images. Six metrics are used for performance evaluation, including accuracy (ACC), sensitivity (SEN), specificity (SPE), F1-Score (F1S), the area under receiver operating characteristic (AUC) and Matthews correlation coefficient (MCC) [12]. Subjects from ADNI-1 are used as the training data, while those from ADNI-2 are treated as independent test data. In \(\text {LM}^3\text {IL}\), 30 landmarks are detected for each MRI via a landmark detection algorithm [9], and these landmarks in each MRI are further located in its corresponding PET image. For each subject, we extract 30 image patches (\( 24\times 24\times 24\)) centered at 30 landmarks from image of each modality (i.e., MRI and PET) as the input of \(\text {LM}^3\text {IL}\).
Our \(\text {LM}^3\text {IL}\) method is compared with five approaches: (1) gray matter (GM) volume within 90 regions-of-interest (denoted as ROI) [5], (2) voxel-wise GM density (denoted as VGD) [13], (3) landmark-based local energy patterns (LLEP) [9], (4) landmark-based deep single-instance learning (LDSIL) [14], and (5) landmark-based deep multi-instance learning (LDMIL) [14] that can be regarded as a single-modal variant of our \(\text {LM}^3\text {IL}\) method using only MRI. To test the effect of our generated PET images, we further compare \(\text {LM}^3\text {IL}\) with its variant (denoted as LM\(^{\mathbf {3}}\)IL-C) that use only subjects with complete MRI and PET data. We share the same landmarks and the same size of image patches in LLEP, LDSIL, LDMIL, \(\text {LM}^3\text {IL}\)-C and \(\text {LM}^3\text {IL}\). Note that four variants of our methods (i.e., LDSIL, LDMIL, \(\text {LM}^3\text {IL}\)-C and \(\text {LM}^3\text {IL}\)) automatically learn features of MRI/PET via deep network, while the remaining methods (ROI, VGD and LLEP) rely on support vector machines with default parameters.
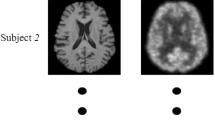
Performance of Image Synthesis Model. To evaluate the quality of the synthetic images generated by 3D-cGAN, we first train the 3D-cGAN model using complete subjects (i.e., containing both PET and MRI) in ADNI-1, and test this image synthesis model on the complete subjects in ADNI-2. Two typical subjects with real and synthetic PET scans are shown in Fig. 4. From Fig. 4, we can observe that our synthetic PET look very similar to their corresponding real images. Also, the mean and standard deviation of PSNR values of synthetic PET images in ADNI-2 are \(24.49\pm 3.46\). These results imply that our trained 3D-cGAN model is reasonable, and the synthetic PET scans have acceptable image quality (in terms of PSNR).
Results of Disease Classification. We further evaluate the effectiveness of our two-stage deep learning method in both tasks of AD classification and MCI conversion prediction. The experimental results achieved by seven different methods are reported in Table 1. From Table 1, we can see that the overall performance of our \(\text {LM}^3\text {IL}\) method is superior to six competing methods regarding six evaluation metrics. Particularly, our method achieves a significantly improved sensitivity value (i.e., nearly 8% higher than the second best sensitivity achieved by ROI) in pMCI vs. sMCI classification. Since higher sensitivity values indicate higher confidence in disease diagnosis, these results imply that our method is reliable in predicting the progression of MCI patients, which is potentially very useful in practice. Besides, as can be seen from Table 1, four methods (i.e., LDSIL, LDMIL, \(\text {LM}^3\text {IL}\)-C and \(\text {LM}^3\text {IL}\)) using deep-learning-based features of MRI and PET usually outperform the remaining three approaches (i.e., ROI, VGD and LLEP) that use hand-crafted features in both classification tasks. This suggests that integrating feature extraction of MRI and PET and classifier model training into a unified framework (as we do in this work) can boost the performance of AD diagnosis. Furthermore, we can see that our \(\text {LM}^3\text {IL}\) method using both MRI and PET generally yields better results than its two single-modal variants (i.e., LDSIL and LDMIL) that use only MRI data. The underlying reason could be that our method can employ the complementary information contained in MRI and PET data. On the other hand, our \(\text {LM}^3\text {IL}\) consistently outperforms \(\text {LM}^3\text {IL}\)-C that utilize only subjects with complete MRI and PET data. These results clearly demonstrate that the synthetic PET images generated by our 3D-cGAN model are useful in promoting brain disease classification performance.
4 Conclusion
In this paper, we have presented a two-stage deep learning framework for AD diagnosis, using incomplete multi-modal imaging data (i.e., MRI and PET). Specifically, in the first stage, to address the issue of missing PET data, we proposed a 3D-cGAN model for imputing those missing PET data based on their corresponding MRI data, considering the relationship between images (i.e., PET and MRI) scanned for the same subject. In the second stage, we developed a landmark-based multi-modal multi-instance neural network for brain disease classification, by using subjects with complete MRI and PET (i.e., both real and synthetic PET). The experimental results demonstrate that the synthetic PET images produced by our method are reasonable, and our proposed two-stage deep learning framework outperforms conventional multi-modal methods for AD classification. Currently, only the synthetic PET images are used for learning the classification models. Using these synthetic MRI data could further augment the training samples for improvement, which will be our future work.
References
Calhoun, V.D., Sui, J.: Multimodal fusion of brain imaging data: a key to finding the missing link(s) in complex mental illness. Biol. Psychiatry: Cogn. Neurosci. Neuroimaging 1(3), 230–244 (2016)
Liu, M., Gao, Y., Yap, P.T., Shen, D.: Multi-hypergraph learning for incomplete multi-modality data. IEEE J. Biomed. Health Inform. 22(4), 1197–1208 (2017)
Parker, R.: Missing Data Problems in Machine Learning. VDM Verlag, Saarbrücken (2010)
Liu, M., Zhang, J., Yap, P.T., Shen, D.: View-aligned hypergraph learning for Alzheimer’s disease diagnosis with incomplete multi-modality data. Med. Image Anal. 36, 123–134 (2017)
Zhang, D., Shen, D.: Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. NeuroImage 59(2), 895–907 (2012)
Zhu, J.Y., Park, T., Isola, P., Efros, A.A.: Unpaired image-to-image translation using cycle-consistent adversarial networks. arXiv preprint arXiv:1703.10593 (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Zhang, J., Gao, Y., Gao, Y.: Detecting anatomical landmarks for fast Alzheimer’s disease diagnosis. IEEE Trans. Med. Imaging 35(12), 2524–2533 (2016)
Boyd, S., Vandenberghe, L.: Convex Optimization. Cambridge University Press, Cambridge (2004)
Jack, C., Bernstein, M., Fox, N.: The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 27(4), 685–691 (2008)
Matthews, B.: Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta (BBA) - Protein Struct. 405(2), 442–451 (1975)
Ashburner, J., Friston, K.J.: Voxel-based morphometry - the methods. NeuroImage 11(6), 805–821 (2000)
Liu, M., Zhang, J., Adeli, E., Shen, D.: Landmark-based deep multi-instance learning for brain disease diagnosis. Med. Image Anal. 43, 157–168 (2018)
Acknowledgment
This research was supported in part by the National Natural Science Foundation of China under Grants 61771397 and 61471297, in part by Innovation Foundation for Doctor Dissertation of NPU under Grants CX201835, and in part by NIH grants EB008374, AG041721, AG042599, EB022880. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and DOD ADNI.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Pan, Y., Liu, M., Lian, C., Zhou, T., Xia, Y., Shen, D. (2018). Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer’s Disease Diagnosis. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11072. Springer, Cham. https://doi.org/10.1007/978-3-030-00931-1_52
Download citation
DOI: https://doi.org/10.1007/978-3-030-00931-1_52
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00930-4
Online ISBN: 978-3-030-00931-1
eBook Packages: Computer ScienceComputer Science (R0)