Abstract
Multi-shell, high resolution diffusion MRI (dMRI) data from the Human Connectome Project (HCP) provides an unprecedented opportunity for the in vivo mapping of human brain pathways. It was recently noted, however, that significant distortions remain present in the data of most subjects preprocessed by the HCP-Pipeline, which have been widely distributed and used extensively in connectomics research. Fundamentally this is caused by the reliance of the HCP tools on the B0 images for registering data from different phase encodings (PEs). In this work, we develop an improved framework to remove the residual distortion in data generated by the HCP-Pipeline. Our method is based on more advanced registration of fiber orientation distribution (FOD) images, which represent information of dMRI scans from all gradient directions and thus provide more reliable contrast to align data from different PEs. In our experiments, we focus on the brainstem area and compare our method with the preprocessing steps in the HCP-Pipeline. We show that our method can provide much improved distortion correction and generate FOD images with more faithful representation of brain pathways.
This work was in part supported by the National Institute of Health (NIH) under Grant R01EB022744, R01AG056573, U01EY025864, P41EB015922.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
With the advance of multiband and several other MRI techniques, the Human Connectome Project (HCP) [1] has developed cutting-edge connectome imaging protocols to acquire high-resolution, multi-shell diffusion MRI (dMRI) that has enabled the in vivo investigation of brain pathways with unprecedented details. Among the various technical advances in the HCP protocol, one notable choice is the acquisition of dMRI data from two phase encodings (PEs) for the correction of susceptibility-induced distortion. However, it was noted recently that majority of the preprocessed dMRI data from HCP [2] still contains significant distortions in regions such as the brainstem [3]. Given the critical role that HCP data continues to play in brain imaging research, it is important to raise awareness of this critical issue in the neuroimaging community and investigate solutions to this fundamental problem.
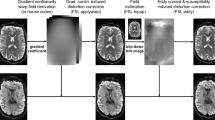
An illustration of the problem in susceptibility distortion correction in HCP-Pipeline. Inputs to the distortion correction based on the topup tool from FSL are the B0 images from the R/L and L/R PE. After that, the corrected B0 images appear “undistorted”. For the ROIs (yellow box) of the corrected data from each PE, we computed the FODs and plotted them in the right column. The residual distortions are clearly visible as highlighted in the dashed ellipsoids. (Color figure online)
The susceptibility of the magnetic field to the tissue/air boundary around the brainstem results in large distortions of the dMRI data along the phase encoding (PE) direction [4]. As shown in Fig. 1, the distortion can be either stretching or compression of the brainstem tissue. The susceptibility-induced distortion not only causes geometric distortion, but more importantly the loss of information due to piling up of signal intensities in regions with severe compression. The state-of-the-art solution is to collect data from opposite PE directions and estimate the distortion field from these two copies of data with the same set of gradient directions [5,6,7], which is the approach HCP and various connectome imaging projects adopt. Using data from the two PEs, the HCP has developed the HCP-Pipeline to correct the distortions and merge them to generate the preprocessed dMRI data for connectivity research [2], which has been widely distributed to the research community. The susceptibility distortion correction in the HCP-Pipeline is mainly based on the topup tool from the FSL, which registers the B0 images from both PEs and estimates the deformation fields for the correction. From the fiber orientation distributions (FODs) [8] plotted in Fig. 1, we can clearly see the brainstem anatomy is severely misaligned even though the “corrected” B0 images appear undistorted. This is because the B0 images do not have enough contrast to resolve the complex brainstem anatomy. Merging such misaligned data, if we continue to run the preprocessing steps in the HCP-Pipeline, will clearly introduce severe artifacts for connectivity modeling. The critical issue is that this is not a rare problem, but prevalent in most preprocessed data [3]. In the benign cases, we might expect distorted pathways. In more serious situations, false pathways can be introduced as we will demonstrate in our experiments. It is thus imperative to tackle this problem and provide improved correction for the highly valuable HCP dataset.
To overcome limitations in current methods for susceptibility distortion correction, we will develop a novel method based on registering the FODs computed from data acquired in each PE. Compared with B0 images which existing methods rely on for distortion correction, FODs represent information from all gradient directions and encode more detailed contrast to allow better alignment of brainstem anatomy. Our method first estimates the FODs for the multi-shell imaging data in each PE. A variational optimization approach will then be developed to minimize the mismatch of FODs from the two PEs while requiring the distortion fields to be opposite transformations as much as possible, which stems from the physical models of susceptibility distortions. After that, we solve a regularized inverse problem to merge these corrected data for connectivity analysis. Compared with previous works based on registering B0 images, there are two main advantages in our FOD-based approach. Because FODs are computed from the ratio between images of each gradient direction to the B0 image, and images from all gradient directions experience the same susceptibility distortion in one acquisition, the Jacobian modulation to image intensities are not needed in our formulation. The second advantage is that there no need of incorporating the rotation operator during the deformation process because the distortion is one-dimensional along the PEs, which leads to a much simplified FOD registration framework. In our experiments, we compare our method with the HCP-Pipeline and demonstrate that much improved distortion correction can be achieved on HCP data.
2 Methods
An overview of the proposed novel method for susceptibility distortion correction is shown in Fig. 2. The inputs to the processing pipeline are the dMRI data from two opposite PEs of the same subject. For data from the HCP, the two PEs are R/L and L/R. In most other connectome imaging studies, the two PEs are typically the A/P and P/A directions. In the first part of the workflow (purple dashed box), the processing steps of the HCP-Pipeline are first applied including the topup and eddy tools from the FSL. In the default HCP-Pipeline, the “corrected” data from both PEs will then be merged to form the output dMRI data. The novel processing pipeline developed in this work is shown in the cyan box in Fig. 2. To remove the residual distortion in the data processed by HCP-Pipeline, we compute the FODs from the multi-shell dMRI data of both PEs to better characterize the differences in these images. The deformation fields from FOD-based registration are then extracted to correct for distortions and merging the data from the two PEs into a final dMRI dataset for connectivity analysis.
2.1 FOD Image Registration
For the multi-shell dMRI data from each PE, we compute the FODs using the multi-compartment model in [8]. At each voxel, the FOD is represented as the coefficients to the spherical harmonics (SPHARMs). For the registration of the FOD images from two PEs, we use the open source software package elastix [9] and solve an image registration optimization problem as follows:
where \(I_F\) and \(I_M\) are the fixed and moving image, respectively, \(\varvec{x}\) is an image voxel location and \(\varvec{T}\left( \varvec{x}, \varvec{\mu }\right) \) is a coordinate transformation parameterized by \(\varvec{\mu }\). In our work, both the fixed and moving images are 4-D images with the 4-th dimension denoting the SPHARM basis functions used for FOD representation. Practically we took the first N SPHARM coefficient images of each PE to form the 4-D vector images for registration. The image mask was generated from the HCP-Pipeline [2]. The mutual information is used as a cost function, and B-spline transformation model is chosen for the modeling of the deformation. An accelerated version of adaptive stochastic gradient descent [10] was used for iterative optimization of Eq. (1). A multi-resolution strategy was used to tackle the local minimum and accelerate the registration procedure. The images were smoothed using a half-reduced Gaussian filter with standard deviations from 32 to 0.5 mm. For each iteration, 10000 image voxels were randomly sampled from the fixed image, and 1500 iterations were used for each resolution.
Following previous works on modeling the susceptibility distortion [5,6,7], we constrained the deformation only along the phase encoding direction during the optimization of the registration. For the HCP data, we denote the FOD from R/L PE as the fixed image, and FOD from L/R PE as the moving image. Using the above registration process, we compute the forward deformation \(\mathbf D \) from R/L to L/R and the inverse deformation \(\text {inv}(\mathbf D )\) from L/R to R/L. For data from opposite PEs, the susceptibility distortion are symmetric in opposite PE direction. For the preprocessed data from HCP-Pipeline, we assume that the residual distortions from the two PEs are still symmetric in each PE direction since the symmetry was modeled in the topup tool [5]. The underlying true, un-distorted image should thus be at the middle point between the fixed and moving image. Half-way deformations \(\mathbf D /2\) and \(\text {inv}(\mathbf D )/2\) will thus be used to merge the data from two PEs and reconstruct the true dMRI image next.
2.2 Merging Data from Different PEs
Using the deformations from FOD-based registration, we will solve an inverse problem to reconstruct the underlying true image \(\varvec{I}\) from two distorted images, denoted as \(\varvec{Y}_{+}\) and \(\varvec{Y}_{-}\), from two different PEs. Because we assume the \(\varvec{I}\) is at the middle point between the fixed \(\varvec{Y}_{+}\) and moving image \(\varvec{Y}_{-}\), we can use the half-way deformations \(\mathbf D /2\) and \(\text {inv}(\mathbf D )/2\) and formulate a forward model as:
where \(K_{+} \) and \(K_{-}\) are sparse matrix representations of the deformations \(\mathbf D /2\) and \(\text {inv}(\mathbf D )/2\) that relates the true image \(\varvec{I}\) to the images from two opposite PEs, and n denotes noise. Using regularized least squares, we can obtain the solution to the inverse problem as
where \(\varvec{R}\) is a smoothness regularization and \(\lambda \) is a non-negative parameter controlling the relative weight of the regularization term. This reconstruction process is applied to data from each gradient direction and form the final output data from our method, which removes the residual distortions and produces high quality data for connectivity analysis.
3 Experimental Results
In this section, we will apply our method to the connectome imaging data from HCP and compare the performance with HCP-Pipeline. In particular, we will focus on the brainstem area, which is a critical region for brain imaging research but usually experiences high distortion. More recently, the brainstem area is considered the earliest site of tau pathology in the Braak staging of Alzheimer’s disease (AD) [11], thus making it highly significant to obtain accurate dMRI imaging data in this challenging area of human brain.
For each HCP subject, we use two raw dMRI scans in the R/L and L/R PEs from the 900-subject release of HCP. Each dMRI scan acquires data from 97 gradient directions distributed on three shells with b-values 1000, 2000, and 3000 \(\text {s}/\text {mm}^2\) at an isotropic spatial resolution of 1.25 mm. For each subject, we will compare the FOD fields with known anatomy in the brainstem to evaluate reconstruction quality. For all experiments, the FODs are calculated with the maximum SPHARM order of 12. The regularization parameter in the merging process is chosen as 0.5. We have conducted our experiments on 10 randomly selected HCP subjects. Overall we observe clear improvements in all subjects after we applied our processing workflows to data generated by HCP-Pipeline. This is even true for subjects that passed the quality control of [3]. Next we show results from two representative subjects to demonstrate the improved distortion corrections achieved by our method.
Results from a brainstem ROI at the level of the midpons of subject 100307. (A) Yellow bar indicates the location of the ROI on the mid-sagittal slice. (B) and (C) show the FODs computed with data from the R/L and L/R PE after preprocessed by HCP-Pipeline, respectively. (D) show the FODs computed with the corrected data from HCP-Pipeline after merging data from R/L and L/R PEs. (E) show the FODs computed with the merged data from our method. (Color figure online)
We first show that residual distortions can still exist in data considered to be high quality. In Fig. 3, we plotted the FODs computed from data generated at different stages of the workflow shown in Fig. 2 for subject 100307, which has passed the quality control in [3] based on visual examination of tract density images (TDI). In both Fig. 3(B) and (C), the dashed ellipsoids highlight the FODs for the left and right cortico-spinal-tract (CST). Clearly they are oriented differently in Fig. 3(B) and (C), which indicates the presence of residual distortions. If we merge these two datasets, as done typically by HCP-Pipeline, and computed the FODs as shown in Fig. 3(D), the FODs corresponding to the left and right CST become mingled together, which is not anatomically correct since the CSTs only decussate at the more inferior medulla area of the brainstem. In the FODs computed from the data generated by our method shown in Fig. 3(E), we highlight that the FODs for the left and right CST in dashed ellipsoids. This shows that our method not only maintains the separation of the two CSTs in midpons, but also corrects the distortions that existed in the data generated by HCP-Pipeline.
In addition to the relatively benign distortions such as in subject 100307, we also observe more critical cases that false fiber trajectories can be caused in the merged data if the distortions are not removed. In Fig. 4, we show the results from subject 136833 at an ROI in the midbrain. In Fig. 4(B) and (C), we show the FODs computed from the data of each PE after they went through the distortion correction steps of HCP-Pipeline. The two dashed ellipsoids highlight the trajectories of the fiber tracts of the cerebellar peduncles on the left and right hemispheres, but they are obviously distorted toward the right and left side, respectively. If we simply merge the data after the distortion correction in HCP-Pipeline, the resulting FODs shown in Fig. 4(D) suggest there are two possible fiber trajectories in each hemisphere as highlighted by the dashed ellipsoid. This is due to the merge of the misaligned data from two PEs and clearly not anatomically meaningful. After we apply the distortion correction by our method, we can see the FOD trajectories of these two fiber pathways become much better aligned and maintains the same number of pathways as in the data from both PEs.
Results from a brainstem ROI at the level of the midbrain of subject 136833. (A) Yellow bar indicates the location of the ROI on the mid-sagittal slice. (B) and (C) show the FODs computed with data from the R/L and L/R PE after preprocessed by HCP-Pipeline, respectively. (D) show the FODs computed with the corrected data from HCP-Pipeline after merging data from R/L and L/R PEs. (E) show the FODs computed with the merged data from our method. (Color figure online)
4 Conclusions and Discussion
There are two main goals of this work. The first is to raise awareness of the residual distortions in the connectome imaging data from HCP and other imaging projects even after they are processed by the HCP-Pipeline. As we showed in our results, these distortions can lead to detrimental effects for modeling the connectivity of brain pathways. The second goal of this work is the development of a novel framework for the correction of these residual distortions in connectome imaging data. Our method is based on the registration of FOD images from both phase encodings, which provides more anatomically relevant contrast than B0 images used in current tools in the HCP-Pipeline. We demonstrated that our method can remove residual distortions and produce anatomically more valid FODs for brainstem regions. Besides the brainstem area, connectome imaging data have distortions throughout the brain, even for deep brain areas around the ventricles. As our future work, it is thus important to conduct a more extensive examination of the residual distortions in other brain regions, and study how our method can help improve the accuracy and reliability in tractography and related connectivity research.
References
Essen, D.V., Ugurbil, K., et al.: The Human Connectome Project: a data acquisition perspective. NeuroImage 62(4), 2222–2231 (2012)
Glasser, M.F., et al.: The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage 80, 105–124 (2013)
Tang, Y., Sun, W., Toga, A.W., Ringman, J.M., Shi, Y.: A probabilistic atlas of human brainstem pathways based on connectome imaging data. NeuroImage 169, 227–239 (2018)
Jezzard, P., Balaban, R.S.: Correction for geometric distortion in echo planar images from B0 field variations. Magn. Reson. Med. 34(1), 65–73 (1995)
Andersson, J.L., Skare, S., Ashburner, J.: How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage 20(2), 870–888 (2003)
Holland, D., Kuperman, J.M., Dale, A.M.: Efficient correction of inhomogeneous static magnetic field-induced distortion in echo planar imaging. NeuroImage 50(1), 175–183 (2010)
Irfanoglu, M.O., Modi, P., Nayak, A., Hutchinson, E.B., Sarlls, J., Pierpaoli, C.: DR-BUDDI (Diffeomorphic Registration for Blip-Up blip-Down Diffusion Imaging) method for correcting echo planar imaging distortions. NeuroImage 106, 284–299 (2015)
Tran, G., Shi, Y.: Fiber orientation and compartment parameter estimation from multi-shell diffusion imaging. IEEE Trans. Med. Imag. 34(11), 2320–2332 (2015)
Klein, S., Staring, M., Murphy, K., Viergever, M.A., Pluim, J.P.: Elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29(1), 196–205 (2010)
Qiao, Y., van Lew, B., Lelieveldt, B.P., Staring, M.: Fast automatic step size estimation for gradient descent optimization of image registration. IEEE Trans. Med. Imaging 35(2), 391–403 (2016)
Braak, H., Thal, D.R., Ghebremedhin, E., Del Tredici, K.: Stages of the pathologic process in Alzheimer disease: age categories from 1 to 100 years. J. Neuropathol. Exp. Neurol. 70(11), 960–969 (2011)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Qiao, Y., Sun, W., Shi, Y. (2018). FOD-Based Registration for Susceptibility Distortion Correction in Connectome Imaging. In: Wu, G., Rekik, I., Schirmer, M., Chung, A., Munsell, B. (eds) Connectomics in NeuroImaging. CNI 2018. Lecture Notes in Computer Science(), vol 11083. Springer, Cham. https://doi.org/10.1007/978-3-030-00755-3_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-00755-3_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00754-6
Online ISBN: 978-3-030-00755-3
eBook Packages: Computer ScienceComputer Science (R0)