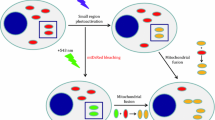
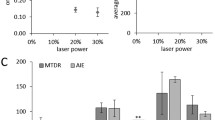
Abstract
Mitochondria are essential organelles of eukaryotic cells with key functions in metabolism, apoptosis, and signaling. As a result, impaired mitochondrial function has been associated with numerous diseases. In order to understand mitochondrial processes, it is fundamental to gain knowledge about their structure and microcompartmentalization, including the function, organization, and dynamics of their protein, nucleic acid, and lipid components. A number of recent groundbreaking advances in fluorescence microscopy enable the study of mitochondrial biology with unprecedented detail. Among them, new methods based on single-molecule and super-resolution microscopy allow us to study mitochondrial structures, protein organizations, and dynamics. Here, we discuss the advantages and disadvantages of different single-molecule microscopy methods to study individual proteins in fixed and living cells in the background of mitochondrial processes, in situ.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Tokunaga M, Imamoto N, Sakata-Sogawa K (2008) Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods 5:159–161
Nyquist H (1928) Certain topics in telegraph transmission theory. Trans Am Inst Electr Eng 47:617–644
Shannon CE (1949) Communication in the presence of noise. Proc IRE 37:10–21
Hess ST, Gould TJ, Gunewardene M, Bewersdorf J, Mason MD (2009) Ultra-high resolution imaging of biomolecules by fluorescence photoactivation localization microscopy (FPALM). Methods Mol Biol 544:483–522
Betzig E et al (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–1645
Hess ST, Girirajan TPK, Mason MD (2006) Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J 91(11):4258–4272
Jungmann R, Steinhauer C, Scheible MB, Kuzyk A, Tinnefeld P, Simmel FC (2010) Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett 10(11):4756–4761
Huang B, Jones SA, Brandenburg B, Zhuang X (2008) Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods 5:1047–1052
Rust MJ, Bates M, Zhuang X (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 3:793–796
Schnitzbauer J et al (2017) Super-resolution microscopy with DNA-PAINT. Nat Protoc 12(6):1198–1228
Kusumi A, Sako Y, Yamamoto M (1993) Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys J 65:2021–2040
Qian H, Sheetz MP, Elson EL (1991) Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys J 60:910–921
Götzke H et al (2019) The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat Commun 10:4403
Nieves DJ et al (2019) tagPAINT: covalent labelling of genetically encoded protein tags for DNA-PAINT imaging. R Soc Open Sci 6(12):191268
Schlichthaerle T et al (2019) Direct visualization of single nuclear pore complex proteins using genetically-encoded probes for DNA-PAINT. Angew Chem Int Ed 58:13004–13008
Heilemann M, Sauer M (2017) Single-molecule localization microscopy in eukaryotes. Chem Rev 117(11):7478–7509
Salvador-Gallego R et al (2016) Bax assembly into rings and arcs in apoptotic mitochondria is linked to membrane pores. EMBO J 35:389–401
Gierlich J et al (2006) Click chemistry as a reliable method for the high-density postsynthetic functionalization of alkyne-modified DNA. Org Lett 8(17):3639–3642
Thorek DLJ, Elias DR, Tsourkas A (2009) Comparative analysis of nanoparticle-antibody conjugations: carbodiimide versus click chemistry. Mol Imaging 8(4):221–229
Agasti SS, Wang Y, Schueder F, Sukumar A, Jungmann R, Yin P (2017) DNA-barcoded labelling probes for highly multiplexed Exchange-PAINT imaging. Chem Sci 8(4):3080–3091. https://doi.org/10.1039/C6SC05420J
Lelek M, Gyparaki MT, Beliu G et al (2021) Single-molecule localization microscopy. Nat Rev Methods Primers 1:39
Stehr F et al (2021) Tracking single particles for hours via continuous DNA-mediated fluorophore exchange. Nat Commun 12:4432
Appelhans T et al (2012) Nanoscale organization of mitochondrial microcompartments revealed by combining tracking and localization microscopy. Nano Lett 12:610–616
Deich J, Judd EM, McAdams HH, Moerner WE (2004) Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. PNAS 101:15921–15926
Gahlmann A, Moerner WE (2014) Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging. Nat Rev Microbiol 12:9–22
Manley S et al (2008) High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods 5:155–157
Schütz GJ, Schindler H, Schmidt T (1997) Single-molecule microscopy on model membranes reveals anomalous diffusion. Biophys J 73:1073–1080
Sergé A, Bertaux N, Rigneault H, Marguet D (2008) Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat Methods 5:687–694
Shen H et al (2017) Single particle tracking: from theory to biophysical applications. Chem Rev 117:7331–7376
Subach FV, Patterson GH, Renz M, Lippincott-Schwartz J, Verkhusha VV (2010) Bright monomeric photoactivatable red fluorescent protein for two-color super-resolution sptPALM of live cells. J Am Chem Soc 132:6481–6491
Kuzmenko A et al (2011) Single molecule tracking fluorescence microscopy in mitochondria reveals highly dynamic but confined movement of Tom40. Sci Rep 1:195
Appelhans T, Busch K (2017) Single molecule tracking and localization of mitochondrial protein complexes in live cells. Methods Mol Biol 1567:273–291
Salewskij K et al (2020) The spatio-temporal organization of mitochondrial F1FO ATP synthase in cristae depends on its activity mode. Biochim Biophys Acta Bioenerg 1861:148091
Pavani SRP et al (2009) Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. PNAS 106:2995–2999
von Diezmann L, Shechtman Y, Moerner WE (2017) Three-dimensional localization of single molecules for super-resolution imaging and single-particle tracking. Chem Rev 117:7244–7275
Beinlich FRM, Drees C, Piehler J, Busch KB (2015) Shuttling of PINK1 between mitochondrial microcompartments resolved by triple-color superresolution microscopy. ACS Chem Biol 10:1970–1976
Appelhans T, Beinlich FRM, Richter CP, Kurre R, Busch KB (2018) Multi-color localization microscopy of single membrane proteins in organelles of live mammalian cells. J Vis Exp 136:57690. https://doi.org/10.3791/57690
Appelhans T, Busch KB (2017) Dynamic imaging of mitochondrial membrane proteins in specific sub-organelle membrane locations. Biophys Rev 9:345–352
Kondadi AK, Anand R et al (2020) Cristae undergo continuous cycles of membrane remodelling in a MICOS-dependent manner. EMBO Rep 21:e49776
Subburaj Y et al (2015) Bax monomers form dimer units in the membrane that further self-assemble into multiple oligomeric species. Nat Commun 6:8042
Cheney PP, Weisgerber AW, Feuerbach AM, Knowles MK (2017) Single lipid molecule dynamics on supported lipid bilayers with membrane curvature. Membranes (Basel) 7:15
Jungmann R et al (2014) Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and exchange-PAINT. Nat Methods 11(3):313–318
Balzarotti F et al (2017) Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 355:606–612
Gwosch KC et al (2020) MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat Methods 17:217–224
Landes CF, Kelly KF, Moringo NA, Tauzin LJ, Hoener BS, Shuang B, Wang W (2016) Super temporal-resolved microscopy (STReM). J Phys Chem Lett 7(22):4524–4529
Wilmes S et al (2015) Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. J Cell Biol 209:579–593
Spahn C et al (2018) Whole-cell, 3D, and multicolor STED imaging with exchangeable fluorophores. Nano Lett 19(1):500–505
Hell SW, Ries J, Ellenberg J, Hoess P, Balzarotti F, Pape JK, Gwosch KC (2020) MINFLUX nanoscopy delivers 3D multicolour nanometer resolution in cells. Nat Methods 17:217–224
Thevathasan JV, Kahnwald M, Cieśliński K, Hoess P, Peneti SK, Reitberger M, Heid D, Kasuba KC, Hoerner SJ, Li Y, Wu Y-L, Mund M, Matti U, Pereira PM, Henriques R, Nijmeijer B, Kueblbeck M, Sabinina VJ, Ellenberg J, Ries J (2019) Nuclear pores as versatile reference standards for quantitative superresolution microscopy. Nat Methods 16(10):1045–1053. https://doi.org/10.1038/s41592-019-0574-9
Ibarra A, Hetzer MW (2015) Nuclear pore proteins and the control of genome functions. Genes Dev 29:337–349
Szymborska A et al (2013) Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341:655–658
Sabinina VJ et al (2021) Three-dimensional superresolution fluorescence microscopy maps the variable molecular architecture of the nuclear pore complex. MBoC 32:1523–1533
Pape JK et al (2020) Multicolor 3D MINFLUX nanoscopy of mitochondrial MICOS proteins. PNAS 117:20607–20614
Eilers Y, Ta H, Gwosch KC, Balzarotti F, Hell SW (2018) MINFLUX monitors rapid molecular jumps with superior spatiotemporal resolution. PNAS 115:6117–6122
Schmidt R et al (2021) MINFLUX nanometer-scale 3D imaging and microsecond-range tracking on a common fluorescence microscope. Nat Commun 12:1478
Huang X, Fan J, Li L et al (2018) Fast, long-term, super-resolution imaging with hessian structured illumination microscopy. Nat Biotechnol 36:451–459
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Dellmann, T., Kostina, A., Garcia Saéz, A.J. (2022). Single-Molecule Microscopy Methods to Study Mitochondrial Processes. In: Šachl, R., Amaro, M. (eds) Fluorescence Spectroscopy and Microscopy in Biology. Springer Series on Fluorescence, vol 20. Springer, Cham. https://doi.org/10.1007/4243_2022_23
Download citation
DOI: https://doi.org/10.1007/4243_2022_23
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-30361-6
Online ISBN: 978-3-031-30362-3
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)