Abstract
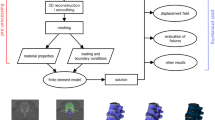
We present a new algorithm for modeling and characteriz- ing shape changes in 3D image sequences of biomedical structures. Our algorithm tracks the shape changes of the objects depicted in the image sequence using an active surface algorithm. To characterize the deformations of the surrounding and inner volume of the object’s surfaces, we use a physics-based model of the objects the image represents. In the applications we are presenting, our physics-based model is linear elasticity and we solve the corresponding equilibrium equations using the Finite Element (FE) method. To generate a FE mesh from the initial 3D image, we have developed a new multiresolution tetrahedral mesh gener- ation algorithm specifically suited for labeled image volumes. The shape changes of the surfaces of the objects are used as boundary conditions to our physics-based FE model and allow us to infer a volumetric deforma- tion field from the surface deformations. Physics-based measures such as stress tensor maps can then be derived from our model for characterizing the shape changes of the objects in the image sequence. Experiments on synthetic images as well as on medical data show the performances of the algorithm.
Chapter PDF
Similar content being viewed by others
References
D. M. Metaxas. Physics-Based Deformable Models: Applications to Computer Vision, Graphics and Medical Imaging. Kluwer Academic Publishers, 1997. 235, 236, 237
D. L. G. Hill, C. R. Maurer, A. J. Martin, S. Sabanathan, W. A. Hall, D. J. Hawkes, D. Rueckert, and C. L. Truwit. Assessment of intraoperative brain deformation using interventional mr imaging. In Berlin Springer-Verlag, editor, MICCAI’ 99, pages 910–919, 1999. 235
M. I. Miga, K. D. Paulsen, P. J. Hoopes, F. E. Kennedy, A. Hartov, and D. W. Roberts. In vivo quantification of a homogeneous brain deformation model for updating preoperative images during surgery. IEEE Transactions on Medical Imaging, 47(2):266–273, February 2000. 235, 236, 245
O. M. Skrinjar and J. S. Duncan. Real time 3d brain shift compensation. In IPMI’ 99, 1999. 235, 236
A. Hagemann, Rohr K., H. S. Stiel, U. Spetzger, and Gilsbach J. M. Biomechanical Modeling of the Human Head for Physically Based, Non-Rigid Image Registration. IEEE Transactions on Medical Imaging, 18(10):875–884, October 1999. 235, 236, 237, 239
Paul M. Thompson, Jay N. Giedd, Roger P. Woods, David MacDonald, Alan C. Evans, and Arthur W. Toga. Growth patterns in the developing brain detected by using continuum mechanical tensor maps. Nature, (404):190–193, 2000. Macmillan Publishers Ltd. 235
H. Delinguette. Toward Realistic Soft-tissue Modeling in Medical Simulation. Proceedings of the IEEE, 86(3):512–523, March 1998. 235, 236
Pengcheng Shi, Albert J. Sinusas, R. Todd Constable, and James S. Duncan. Volumetric deformation using mechanics-based data fusion: Applications in cardiac motion recovery. International Journal of Computer Vision, November 1999. 235
J. Martin, A. Pentland, S. Sclaro., and R. Kikinis. Characterization of Neuropathological Shape Deformations. IEEE Transactions on Pattern Analysis and Machine Intelligence, 20(2):97–112, February 1998. 236
Jean-Philippe Thirion and Guillaume Calmon. Deformation analysis to detect and quantify active lesions in 3d medical image sequences. Technical Report 3101, INRIA Sophia Antipolis, February 1997. 236
Stephen M. Pizer, Christina A. Burbeck, Daniel S. Fritch, Bryan S. Morse, Alan Liu, Shobha Murthy, and Derek T. Pu.. Human Perception and Computer Image Analysis of Objects in Images. In DICTA-93 Digital Image Computing: Techniques and Applications, volume 1, pages 19–26, 1993. 236
G. Stetten and S. M. Pizer. Medial Node Models to identify and Measure Objects in Real-Time 3D Echocardiography. IEEE Transactions on Medical Imaging, 18(10):1025–1034, 1999. 236
S. M. Pizer, D. S. Fritsch, P. Yushkevich, V. Johnson, and E. Chaney. Segmentation, registration, and measurement of shape variation via image object shape. IEEE Transactions on Medical Imaging, pages 851–865, 1996. 236
L. D. Cohen and Cohen I. Finite Element Methods for Active Contour Models and Balloons for 2D and 3D Images. IEEE Transactions on Pattern Analysis and Machine Intelligence, 15:1131–1147, 1993. 236, 242
C. Davatzikos. Spatial Transformation and Registration of Brain Images Using Elastically Deformable Models. Computer Vision and Image Understanding, 66(2):207–222, May 1997. 236
P. Thompson and A. W. Toga. A Surface-Based Technique for Warping Three-Dimensional Images of the Brain. IEEE Transactions on Medical Imaging, 15(4):402–417, 1996. 236
R. Bajcsy and S. Kovacic. Multi-resolution Elastic Matching. Computer Vision, Graphics, and Image Processing, 46:1–21, 1989. 236
G. E. Christensen, S. C. Joshi, and M. I. Miller. Volumetric Transformation of Brain Anatomy. IEEE Transactions on Medical Imaging, 16(6):864–877, December 1997. 236
M. Bro-Nielsen and C. Gramkow. Fast Fluid Registration of Medical Images. In Visualization in Biomedical Computing (VBC’ 96), pages 267–276, 1996. 236
X. Papademetris, A. J. Sinusas, D. P. Dione, and J. S. Duncan. 3D Cardiac Deformation from Ultrasound Images. In MICCAI 1999: Medical Image Computing and Computer Assisted Intervention, pages 420–429. Springer, September 1999. 236
J. Park, D. Metaxas, and L. Axel. Analysis of left ventricular wall motion based on volumetric deformable models and MRI-SPAMM. Medical Image Analysis, 1(1):53–71, 1996. 236
E. Haber, D. N. Metaxas, and L. Axel. Motion Analysis of the Right Ventricle from MR images. In MICCAI 1998: Medical Image Computing and Computer Assisted Intervention, pages 177–188. Springer, October 1998. 236
S. K. Kyriacou, C. Davatzikos, S. J. Zinreich, and R. N. Bryan. Nonlinear elastic registration of brain images with tumor pathology using a biomechanical model. IEEE Transactions on Medical Imaging, 18(7):580–592, july 1999. 236
O. C. Zienkewickz and R. L. Taylor. The Finite Element Method. McGraw Hill Book Co., 1987. 238, 239
Will Schroeder, Ken Martin, and Bill Lorensen. The Visualization Toolkit: An Object-Oriented Approach to 3D Graphics. Prentice Hall PTR, New Jersey, second edition, 1998. 239
B. Geiger. Three dimensional modeling of human organs and its application to diagnosis and surgical planning. Technical Report 2105, INRIA, 1993. 239
M. Kass, A. Witkin, and D. Terzopoulos. Snakes: Active Contour Models. International Journal of Computer Vision, 1(4):321–331, 1988. 242
O. Cuisenaire and B. Macq. Fast Euclidean Distance Transformation by Propagation Using Multiple Neighborhoods. Computer Vision and Image Understanding, 76(2):163–172, November 1999. 242
M. Ferrant, O. Cuisenaire, and B. Macq. Multi-Object Segmentation of Brain Structures in 3D MRI Using a Computerized Atlas. In SPIE Medical Imaging’ 99, volume 3661-2, pages 986–995, 1999. 243
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2000 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Ferrant, M., Macq, B., Nabavi, A., Warfield, S.K. (2000). Deformable Modeling for Characterizing Biomedical Shape Changes. In: Borgefors, G., Nyström, I., di Baja, G.S. (eds) Discrete Geometry for Computer Imagery. DGCI 2000. Lecture Notes in Computer Science, vol 1953. Springer, Berlin, Heidelberg. https://doi.org/10.1007/3-540-44438-6_20
Download citation
DOI: https://doi.org/10.1007/3-540-44438-6_20
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-41396-7
Online ISBN: 978-3-540-44438-1
eBook Packages: Springer Book Archive