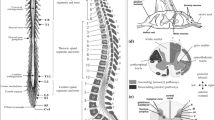
Abstract
Spinal cord analysis is an important problem relating to the study of various neurological diseases. We present a novel approach to spinal cord segmentation in magnetic resonance images. Our method uses 3D “deformable organisms” (DefOrg) an artificial life framework for medical image analysis that complements classical deformable models (snakes and deformable meshes) with high-level, anatomically-driven control mechanisms. The DefOrg framework allows us to model the organism’s body as a growing generalized tubular spring-mass system with an adaptive and predominantly elliptical cross section, and to equip them with spinal cord specific sensory modules, behavioral routines and decision making strategies. The result is a new breed of robust DefOrgs, “spinal crawlers”, that crawl along spinal cords in 3D images, accurately segmenting boundaries, and providing sophisticated, clinically-relevant structural analysis. We validate our method through the segmentation of spinal cords in clinical data and provide comparisons to other segmentation techniques.
Chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
References
Mathias, J., Tofts, P., Losseff, N.: Texture analysis of spinal cord pathology in multiple sclerosis. Magnetic Resonance in Medicine 42, 929–935 (1999)
D’Haese, P., Niermann, K., Cmelak, A., Duay, V., Li, R., Dawant, B.: Atlas-based spine segmentation for radiotherapy planning. American Society of Therapeutic Radiology and Oncology (2004)
Mancas, M., Gosselin, B., Macq, B.: Segmentation using a region-growing thresholding. In: Proceedings of the SPIE, vol. 5672, pp. 388–398 (2005)
Karangelis, G., Zimeras, S.: An accurate 3d segmentation method of the spinal canal applied to CT data. In: Bildverarbeitung fur die Medizin, pp. 370–373 (2002)
Schmit, B.D., Cole, M.K.: Quantification of morphological changes in the spinal cord in chronic human spinal cord injury using magnetic resonance imaging. IEEE EMBS 26, 4425–4428 (2004)
Coulon, O., Hickman, S.J., Parker, G., Barker, G., Miller, D., Arridge, S.: Quantification of spinal cord atrophy from magnetic resonance images via a b-spline active surface model. Magnetic Resonance in Medicine 47, 1176–1185 (2002)
Aylward, S., Bullit, E., Pizer, S., Eberly, D.: Intensity ridge and widths for tubular object segmentation and description. In: IEEE/SIAM workshop Mathematical Methods Biomedical Image Analysis, pp. 131–138 (1996)
McIntosh, C., Hamarnerh, G.: Vessel crawlers: 3d physically-based deformable organisms for vasulature segmentation and analysis. In: IEEE Conference on Computer Vision and Pattern Recognition, vol. 1, pp. 1084–1091 (2006)
Hamarneh, G., McInerney, T., Terzopoulos, D.: Deformable organisms for automatic medical image analysis. In: Niessen, W.J., Viergever, M.A. (eds.) MICCAI 2001. LNCS, vol. 2208, pp. 66–76. Springer, Heidelberg (2001)
Lindeberg, T.: Scale-Space Theory in Computer Vision. Kluwer Academic Publishers, Norwell (1994)
Frangi, A.F., Niessen, W.J., Vincken, K.L., Viergever, M.A.: Multiscale vessel enhancement filtering. In: Wells, W.M., Colchester, A.C.F., Delp, S.L. (eds.) MICCAI 1998. LNCS, vol. 1496, pp. 130–137. Springer, Heidelberg (1998)
Hamarneh, G., McIntosh, C.: Physics-based deformable organisms for medical image analysis. In: SPIE Medical Imaging, vol. 5747, pp. 326–335 (2005)
Yushkevich, P.A., Piven, J., Hazlett, H.C., Smith, R.G., Ho, S., Gee, J.C., Gerig, G.: User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage (2006)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2006 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
McIntosh, C., Hamarneh, G. (2006). Spinal Crawlers: Deformable Organisms for Spinal Cord Segmentation and Analysis. In: Larsen, R., Nielsen, M., Sporring, J. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2006. MICCAI 2006. Lecture Notes in Computer Science, vol 4190. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11866565_99
Download citation
DOI: https://doi.org/10.1007/11866565_99
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-44707-8
Online ISBN: 978-3-540-44708-5
eBook Packages: Computer ScienceComputer Science (R0)