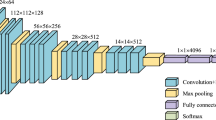
Abstract
Cell classification by computer assistant technique has become a popular topic recently. It is crucial to do early disease diagnosis with less cost. In order to find efficient, high accuracy computer assistant methods, The International Symposium on Biomedical Imaging (ISBI) held the classification of normal vs malignant cells (C-NMC) challenge 2019 for classifying B-ALL white blood cancer microscopic images. Our team deploys several state-of-the-art CNN architectures and uses an ensemble method named stacking to boost the final prediction performance, which helped to get the eighth place in the preliminary phase and tenth place in the final phase. Weighted F1 scores evaluated on our model were 0.8674 and 0.8552, for the preliminary and final phase, respectively. It is a relatively reasonable result in such an expertise required task.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Pui, C.H.: Acute lymphoblastic leukemia. Pediatr. Clin. North Am. 44(4), 831 (1997)
Nazlibilek, S., Karacor, D., Ercan, T., et al.: Automatic segmentation, counting, size determination and classification of white blood cells. Measurement 55, 58–65 (2014)
Wang, D., Khosla, A., Gargeya, R., et al.: Deep learning for identifying metastatic breast cancer (2016)
Grewal, M., Srivastava, M.M., Kumar, P., et al.: RADNET: radiologist level accuracy using deep learning for HEMORRHAGE detection in CT scans (2017)
Esteva, A., Kuprel, B., Novoa, R.A., et al.: Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639), 115–118 (2017)
Kamnitsas, K., Ledig, C., Newcombe, V.F.J., et al.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. (2016). S1361841516301839
Voets, M., Møllersen, K., Bongo, L.A.: Replication study: development and validation of deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs (2018)
Rajpurkar, P., Hannun, A.Y., Haghpanahi, M., et al.: Cardiologist-level Arrhythmia detection with convolutional neural networks (2017)
Duggal, R., Gupta, A., Gupta, R., Wadhwa, M., Ahuja, C.: Overlapping cell nuclei segmentation in microscopic images using deep belief networks. In: Indian Conference on Computer Vision, Graphics and Image Processing (ICVGIP), India, December 2016
Duggal, R., Gupta, A., Gupta, R.: Segmentation of overlapping/touching white blood cell nuclei using artificial neural networks. CME Series on Hemato-Oncopathology, All India Institute of Medical Sciences (AIIMS), New Delhi, India, July 2016
Duggal, R., Gupta, A., Gupta, R., Mallick, P.: SD-layer: stain deconvolutional layer for CNNs in medical microscopic imaging. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds.) Medical Image Computing and Computer-Assisted Intervention, MICCAI 2017. Lecture Notes in Computer Science, Part III, LNCS 10435, pp. 435–443. Springer, Cham. https://doi.org/10.1007/978-3-319-66179-7_50
Deng, J., Dong, W., Socher, R., et al.: ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE (2009)
Rajpurkar, P., Irvin, J., Zhu, K., et al.: CheXNet: radiologist-level pneumonia detection on chest X-rays with deep learning (2017)
Deng, L., Yu, D.: Deep learning: methods and applications. Found. Trends Signal Process. 7(3), 197–387 (2014)
Gupta, A., Duggal, R., Gupta, R., Kumar, L., Thakkar, N., Satpathy, D.: GCTI-SN: Geometry-Inspired Chemical and Tissue Invariant Stain Normalization of Microscopic Medical Images. Under review
Gupta, R., Mallick, P., Duggal, R., Gupta, A., Sharma, O.: Stain color normalization and segmentation of plasma cells in microscopic images as a prelude to development of computer assisted automated disease diagnostic tool in multiple Myeloma. In: 16th International Myeloma Workshop (IMW), India, March 2017
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: CVPR (2016)
Huang, G., Liu, Z., Weinberger, K.Q., Maaten, L.: Densely connected convolutional networks. In: CVPR (2017)
Szegedy, C., Ioffe, S., Vanhoucke, V.: Inception-v4, Inception-ResNet and the impact of residual connections on learning. In: CoRR (2016). arXiv:1602.07261
Breiman, L.: Stacked regressions. Mach. Learn. 24(1), 49–64 (1996)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Ding, Y., Yang, Y., Cui, Y. (2019). Deep Learning for Classifying of White Blood Cancer. In: Gupta, A., Gupta, R. (eds) ISBI 2019 C-NMC Challenge: Classification in Cancer Cell Imaging. Lecture Notes in Bioengineering. Springer, Singapore. https://doi.org/10.1007/978-981-15-0798-4_4
Download citation
DOI: https://doi.org/10.1007/978-981-15-0798-4_4
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-0797-7
Online ISBN: 978-981-15-0798-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)