Abstract
During the last decade pathology has benefited from the rapid progress of image digitizing technologies, which led to the development of scanners, capable to produce so-called Whole Slide images (WSI) which can be explored by a pathologist on a computer screen comparable to the conventional microscope and can be used for diagnostics, research, archiving and also education and training. Digital pathology is not just the transformation of the classical microscopic analysis of histological slides by pathologists to just a digital visualization. It is a disruptive innovation that will dramatically change medical work-flows in the coming years and help to foster personalized medicine. Really powerful gets a pathologist if she/he is augmented by machine learning, e.g. by support vector machines, random forests and deep learning. The ultimate benefit of digital pathology is to enable to learn, to extract knowledge and to make predictions from a combination of heterogenous data, i.e. the histological image, the patient history and the *omics data. These challenges call for integrated/integrative machine learning approach fostering transparency, trust, acceptance and the ability to explain step-by-step why a decision has been made.
Similar content being viewed by others
Keywords
1 Introduction and Motivation
The ability to mine “sub-visual” image features from digital pathology slide images, features that may not be visually discernible by a pathologist, offers the opportunity for better quantitative modeling of disease appearance and hence possibly improved prediction of disease aggressiveness and patient outcome. However, the compelling opportunities in precision medicine offered by big digital pathology data come with their own set of computational challenges. Image analysis and computer assisted detection and diagnosis tools previously developed in the context of radiographic images are woefully inadequate to deal with the data density in high resolution digitized whole slide images. Additionally, there has been recent substantial interest in combining and fusing radiologic imaging, along with proteomics and genomics based measurements with features extracted from digital pathology images for better prognostic prediction of disease aggressiveness and patient outcome. Again there is a paucity of powerful tools for combining disease specific features that manifest across multiple different length scales. The purpose of this paper is to discuss developments in computational image analysis tools for predictive modeling of digital pathology images from a detection, segmentation, feature extraction, and tissue classification perspective. We discuss the emergence of new handcrafted feature approaches for improved predictive modeling of tissue appearance and also review the emergence of deep learning schemes for both object detection and tissue classification. We also briefly review some of the state of the art in fusion of radiology and pathology images and also combining digital pathology derived image measurements with molecular “omics” features for better predictive modeling [1].
The adoption of data-intensive methods can be found throughout various branches of health, leading e.g. to more evidence-based decision-making and to help to go towards personalized medicine [2]: A grand goal of future biomedicine is to tailor decisions, practices and therapies to the individual patient. Whilst personalized medicine is the ultimate goal, stratified medicine has been the current approach, which aims to select the best therapy for groups of patients who share common biological characteristics. Here, ML approaches are indispensable, for example causal inference trees (CIT) and aggregated grouping, seeking strategies for deploying such stratified approaches. Deeper insight of personalized treatment can be gained by studying the personal treatment effects with ensemble CITs [3]. Here the increasing amount of heterogenous data sets, in particular “-omics” data, for example from genomics, proteomics, metabolomics, etc. [4] make traditional data analysis problematic and optimization of knowledge discovery tools imperative [5, 6]. On the other hand, many large data sets are indeed large collections of small data sets. This is particularly the case in personalized medicine where there might be a large amount of data, but there is still a relatively small amount of data for each patient available [7]. Consequently, in order to customize predictions for each individual it is necessary to build a model for each patient along with the inherent uncertainties, and to couple these models together in a hierarchy so that information can be “borrowed” from other similar patients. This is called model personalization, and is naturally implemented by using hierarchical Bayesian approaches including e.g. hierarchical Dirichlet processes [8] or Bayesian multi-task learning [9].
This variety of problems in Digital Pathology requires a synergistic combination of various methodological approaches which calls for a combination of various approaches, e.g. geometrical approaches with deep learning models [10].
After a short motivation and explanation of why machine aided pathology is interesting, relevant and important for the future of diagnostic medicine, this article is organized as follows:
In Sect. 2 we provide a glossary of the most important terms.
In Sect. 3 we give an overview of where digital pathology is already in use today, which technologies of slide scanning are currently state-of-the-art, and describe the next steps towards a machine aided pathology. A sample use-case shall demonstrate the typical work-flow. Because data-integration, data fusion and data-preprocessing is an important aspect, we briefly describe these issues here.
In Sect. 4 we describe the most promising state-of-the-art machine learning technologies which can be of use for digital pathology.
In Sect. 5, finally, we discuss some important future challenges in machine learning, which includes multi-task learning, transfer learning and the use of multi-agent-hybrid systems.
2 Glossary and Key Terms
Automatic Machine Learning (aML) in bringing the human-out-of-the-loop is the grand goal of ML and works well in many cases having “big data” [11].
Big Data is indicating the flood of data today; however, large data sets are necessary for aML approaches to learn effectively; the problem is in “dirty data” [12], and sometimes we have large collections of little, but complex data.
Data Fusion is the process of integration multiple data representing the same real-world object into one consistent, accurate, and useful representation.
Data Integration is combining data from different sources and providing a unified view.
Deep Learning allows models consisting of multiple layers to learn representations of data with multiple levels of abstraction [13].
Digital Pathology is not only the conversion of histopathological slides into a digital image (WSI) that can be uploaded to a computer for storage and viewing, but a complete new medical work procedure.
Dimensionality of data is high, when the number of features p is larger than the number of observations n by magnitudes. A good example for high dimensional data is gene expression study data [14].
Explainability is motivated due to lacking transparency of black-box approaches, which do not foster trust and acceptance of ML among end-users. Rising legal and privacy aspects, e.g. with the new European General Data Protection Regulations, make black-box approaches difficult to use, because they often are not able to explain why a decision has been made [15].
interactive Machine Learning (iML) in bringing the human-in-the-loop is beneficial when having small amounts of data (“little data”), rare events or dealing with complex problems [16, 17], or need reenactment (see explainability).
Knowledge Discovery (KDD) includes exploratory analysis and modeling of data and the organized process to identify valid, novel, useful and understandable patterns from these data sets [18].
Machine Aided Pathology is the management, discovery and extraction of knowledge from a virtual case, driven by advances of digital pathology supported by feature detection and classification algorithms.
Multi-Task Learning (MTL) aims to learn a problem together with multiple, different but related other problems through shared parameters or a shared representation. The underlying principle is bias learning based on probable approximately correct learning (PAC learning) [19].
Topological Data Mining uses algebraic geometry to recover parameters of mixtures of high-dimensional Gaussian distributions [20].
Transfer Learning How can machine learning algorithms perform a task by exploiting knowledge, extracted during solving previous tasks? Contributions to solve this problem would have major impact to Artificial Intelligence generally, and Machine Learning specifically [21].
Virtual Case is the set of all histopathological slides of a case together with meta data from the macro pathological diagnosis [22].
Virtual Patient has very different definitions (see [23], we define it as a model of electronic records (images, reports, *omics) for studying e.g. diseases.
Visualization can be defined as transforming the symbolic into the geometric and the graphical presentation of information, with the goal of providing the viewer with a qualitative understanding of the information contents [6, 24].
Whole Slide Imaging (WSI) includes scanning of all tissue covered areas of a histopathological slide in a series of magnification levels and optional as a set of focus layers.
3 From Digital Pathology to Machine Aided Pathology
3.1 Digital Pathology
Modern pathology was founded by Rudolf Virchow (1821-1902) in the mid of the \(19^{\text {th}}\) century. In his collection of lectures on Cellular Pathology (1858) he set the basis of modern medical science and established the “microscopically thinking” still applied today by every pathologist. In histopathology a biopsy or surgical specimen is examined by a pathologist, after the specimen has been processed and histological sections have been placed onto glass slides. In cytopathology either free cells (fluids) or tissue micro-fragments are “smeared” on a slide without cutting a tissue.
In the end of the \(20^{\text {th}}\) century an individual clinical pathologist was no longer able to cover the knowledge of the whole scientific field. This led to today’s specialization of clinical pathology either by organ systems or methodologies. Molecular biology and *omics technologies set the foundation for the emerging field of molecular pathology, which today alongside WSI provides the most important source of information, especially in the diagnosis of cancer and infectious diseases.
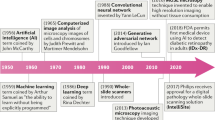
The roots of digital pathology go back to the 1960s, when first telepathology experiments took place. Later in the 1990 s the principle of virtual microscopy [25] appeared in several life science research areas. At the turn of the century the scientific community more and more agreed on the term “digital pathology” [26] to denote digitization efforts in pathology.
However in 2000 the technical requirements (scanner, storage, network) were still a limiting factor for a broad dissemination of digital pathology concepts. Over the last 5 years this changed as new powerful and affordable scanner technology as well as mass/cloud storage technologies appeared on the market. This is also clearly reflected in the growing number of publications mentioning the term “digital pathology” in PMC, see Fig. 1.
The field of Radiology has undergone the digital transformation almost 15 years ago, not because radiology is more advanced, but there are fundamental differences between digital images in radiology and digital pathology: The image source in radiology is the (alive) patient, and today in most cases the image is even primarily captured in digital format. In pathology the scanning is done from preserved and processed specimens, for retrospective studies even from slides stored in a biobank. Besides this difference in pre-analytics and metadata content, the required storage in digital pathology is two to three orders of magnitude higher than in radiology. However, the advantages anticipated through digital pathology are similar to those in radiology:
-
Capability to transmit digital slides over distances quickly, which enables telepathology scenarios.
-
Capability to access past specimen from the same patients and/or similar cases for comparison and review, with much less effort then retrieving slides from the archive shelves.
-
Capability to compare different areas of multiple slides simultaneously (slide by slide mode) with the help of a virtual microscope.
-
Capability to annotate areas directly in the slide and share this for teaching and research.
Digital pathology is today widely used for educational purposes [27] in telepathology and teleconsultation as well as in research projects. Digital pathology in diagnostics is an emerging and upcoming field. With the validation of the first WSI systems for primary diagnosis by the FDA the first steps for the digital transition in pathology are done, and we anticipate a major paradigm shift within the next 10 years.
Sharing the perception of the instructor when looking through a microscope is a technical challenge. Digital pathology allows to share and annotate slides in a much easier way. Also the possibility to download annotated lecture sets generates new opportunities for e-learning and knowledge sharing in pathology.
The level of specialization in pathology is ever increasing, and it is no more possible to cover at small and medium size pathology institutes all fields, so expert knowledge is often missing to generate the best possible diagnosis for the patient. This is a main driving force for telepathology, when a local team can easily involve specialists that don’t need to be in the same location, and/or get a specialized second opinion. For overnight diagnosis workflows even the time difference between different countries can be utilized, e.g. the diagnosis for a virtual case scanned in France in the evening can be ready next morning, done by a pathologist in Canada.
It is important that in all use cases the digital slides are archived in addition to the analogue tissue blocks and slides. This will (a) ensure documentation and reproducibility of the diagnosis (an additional scan will never produce the same WSI) and (b) generate a common and shared pool of virtual cases for training and evaluation of machine learning algorithms. Archiving WSI is even a prerequisite for the validation and documentation of diagnostic workflows, especially when algorithmic quantification and classification algorithms are applied. In the next sections we describe requirements for data management and digital slide archiving as a starting point for machine aided pathology scenarios.
3.2 Virtual Case
A pathological workflow always starts with the gross evaluation of the primary sample. Depending on the medical question and the material type small tissue parts are extracted from the primary sample and are either embedded in a paraffin block or cryo-frozen. From the tissue blocks the pathology labs cuts several slides, applies different staining methods and conducts additional histological and molecular tests. Finally, the pathologists evaluate all the slides together with the supporting gross-and molecular findings and makes the diagnosis. If in addition to the set of WSI all information is present in a structured digital format, we call this a virtual case. In a virtual case, the average number of slides and additional findings varies very much for different medical questions and material types. Figure 2 shows the average number of slides for different diagnosis done in the year 2016 at the Institute of Pathology at Graz Medical University.
The most demanding data elements in a virtual case are the whole slide images (WSI). Compared to radiology, where the typical file size are between 131 KB for MRI images, 524 KB for CT-Scans, 18 MB for digital radiology, 27 MB for digital mammography and 30 MB for computed radiography [28], a single WSI scan with 80x magnification consists of 15.6 Gigapixels. For the calculation of the WSI file size and comparison of different scanner manufacturers, we use the de-facto standard area of 15 mm x 15 mm, with an optical resolution of 0.12 \({\upmu }\)m, which corresponds to an 80x magnification (see Fig. 3).
With 8 bit information for each color channel a WSI results in 46.9 GB stored in an uncompressed image format. Looking at the number of slides of a typical case, it is clear, that some compression techniques must be applied to the image data, and luckily several studies reported that lossy compression with a high quality level does not influence the diagnostic results. Still there are unresolved questions:
-
High compression levels. Can the compression level be increased up to 200 without significant influence in human decision making, e.g. with optimized jpeg2000 algorithms and intra-frame compression techniques for z-layers.
-
Tissue/Staining dependencies. Does the maximum compression level depend on tissue type and staining?
-
Compression in ML scenarios. What compression level can be applied when WSI images are part of machine learning training sets and/or classified by algorithms?
The newest generation of scanners (as of 2017 !) is able to digitize a slide at various vertical focal planes, called z-layers, each the size of a singe layer. The multi-layer image can be either combined by algorithms to a single composite multi-focus image (Z-stacking) or used to simulate the fine focus control of a conventional microscope. Z-stacking is a desirable feature especially when viewing cytology slides, however, the pathologist should be aware that such an image can never be seen through the microscope (see Fig. 4).
At the Institute of Pathology at Graz Medical University, which is a medium to large organization, about 73,000 diagnosis are made within a year and approx 335,000 glass slides are produced in the pathology lab, approx 25,000 glass slides in the cytology lab. This results in a yearly storage capacity of almost 1 PetaByte and the appropriate computing power to process approx. 1000 slides per day plus the necessary capacity to train and improve ML algorithms. This numbers illustrate that the digital transformation of diagnostic workflows in pathology will demand for very high storage, even when stored in a compressed format, as well as computing capacity.
Several data formats are used today, either vendor independent (DICOM, TIFF/BigTIFF, Deep Zoom images) and vendor specific formats from Aperio, Hamamatsu, Leica, 3DHistech, Philips, Sakura and Trestle. In the setup of a virtual slide archive for medical research and machine learning it is essential to (a) agree on a common exchange format, and (b) to separate patient related and image related metadata. Patient related metadata comprise direct identifiers (name, birthday, zip code, ...) but also diagnosis results and others results from the patient medical history. When no such data is stored within or attached to the image format, the WSI is purely anonymous, as no re-identification of the patient is possible. To link between the same WSI used in different studies, either a global unique identifier (GUID) or a image generated hash can be used.
3.3 Towards Machine Aided Pathology
Digitizing workflows is one important enabling step to a groundbreaking change in clinical pathology, where AI methods and ML paradigms are introduced to pathological diagnosing. This assistance starts with simple classification and quantification algorithms as already available today, and ends in a full autonomous pathologist, where human expertise is replaced by machine intelligence. To distinguish such scenarios from simple digital workflows we propose the term machine aided pathology, when a significant contribution of the decision making process is supported by machine intelligence. Machine aided pathology solutions can be applied at several steps of the diagnosis making process:
-
Formulation of a hypothesis. Each diagnosis starts with a medical question and a corresponding underlying initial hypothesis. The pathologist refines this hypothesis in an iterative process, consequently looking for known patterns in a systematic way in order to confirm, extend or reject his/her initial hypothesis. Unconsciously, the pathologist asks the question “What is relevant?” and zooms purposefully into the -according to his/her opinion - essential areas of the cuts. The duration and the error rate in this step vary greatly between inexperienced and experienced pathologists. An algorithmic support in this first step would contribute in particular to the quality and interoperability of pathological diagnoses and reduce errors at this stage, and would be particularly helpful for educational purposes. A useful approach is known from Reeder and Felson (1975) [29] to recognize so called gamuts in images and to interpret these according to the most likely and most unlikely, an approach having its origin in differential diagnosis.
-
–
Very large amounts of data can only be managed with a “multi resolution” image processing approach using image pyramids. For example, a Colon cancer case consists of approximately 20 Tera (!) pixel of data - a size which no human is capable of processing.
-
–
The result of this complex process is a central hypothesis, which has to be tested on a selection of relevant areas in the WSI, which is determined by quantifiable values (receptor status, growth rate, etc.).
-
–
Training data sets for ML can now contain human learning strategies (transfer learning) as well as quantitative results (hypotheses, areas, questions, etc.).
-
–
-
Detection and classification of known features. Through a precise classification and quantification of selected areas in the sections, the central hypothesis is either clearly confirmed or rejected. In this case, the pathologist has to consider that the entire information of the sections is no longer taken into account, but only areas relevant to the decision are involved. It is also quite possible that one goes back to the initial hypothesis step by step and changes their strategy or consults another expert, if no statement can be made on the basis of the classifications.
-
–
In this step ML algorithms consist of well known standard classification and quantification approaches.
-
–
An open question is how to automatically or at least semi-automatically produce training sets, because here specific annotations are needed (which could come from a stochastic ontology, e.g.).
-
–
Another very interesting and important research question is, whether and to what extent solutions learned from one tissue type (organ 1) can be transferred to another tissue type (organ 2) – transfer learning – and how robust the algorithms are with respect to various pre-analytic methods, e.g. stainings, etc.
-
–
-
Risk prediction and identification of unknown features. Within the third step, recognized features (learned parameters) are combined to a diagnosis and an overall prediction of survival risk. The main challenge in this step lies in training/validation and in the identification of novel, previously unknown features from step two. We hypothesize that the pathologist supported by machine learning approaches is able to discover patterns – which previously were not accessible! This would lead to new insights into previously unseen or unrecognized relationships.
Besides challenges in ML, also the following general topics and prerequisites have to be solved for a successful introduction of machine aided pathology:
-
Standardization of WSI image formats and harmonization of annotation/metadata formats. This is essential for telepathology applications and even more important for the generation of training sets, as for a specific organ and disease stages, even at a large institute of pathology the required amount of cases may not be available.
-
Common digital cockpit and visualization techniques should be used in education, training and across different institutes. Changing the workplace should be as easy as switching the microscope model or manufacturer. However, commonly agreed-upon visualization and interaction paradigms can only be achieved in a cross vendor approach and with the involvement of the major professional associations.
3.4 Data Integration
The image data (see Fig. 5) can be fused with two other sources of data: (1) Clinical data from electronic patient records [30], which contain documentations, reports, but also laboratory tests, physiological parameters, recorded signals, ECG, EEG, etc.); this also enables linking to other image data (standard X-ray, MR, CT, PET, SPECT, microscopy, confocal laser scans, ultrasound imaging, molecular imaging, etc.) (2) *omics data [4], e.g. from genomic sequencing technologies (Next Generation Sequencing, NGS, etc.), microarrays, transcriptomic technologies, proteomic and metabolomic technologies, etc., which all plays important roles for biomarker discovery and drug design [31, 32].
Detail of a typical WSI: Hematoxylin and eosinstained histological section of a formalin-fixed and paraffin-embedded normal human liver tissue. Manual annotation: PV, portal vein; BD, bilde duct; HA, hepatic artery, HC (arrow), example of hepatocyte. Bar = 30 \(\upmu \)m
Data integration is a hot topic in health informatics generally and solutions can bridge the gap between clinical routine and biomedical research [33]. This is becoming even more important due to the heterogeneous and different data sources, including picture archiving and communication systems (PACS) and radiological information systems (RIS), hospital information systems (HIS), laboratory information systems (LIS), physiological and clinical data repositories, and all sorts of -omics data from laboratories, using samples from biobanks. Technically, data integration is the combination of data from different sources and providing users with a unified view on these data, whereas data fusion is matching various data sets representing one and the same object into a single, consistent and clean representation [34]; in health informatics these unified views are particularly important in high-dimensions, e.g. for integrating heterogeneous descriptions of the same set of genes [35]. The general rule is that fused data is more informative than the original separate inputs. Inclusion of these different data sources and a fresh look on the combined views would open future research avenues [36].
4 Machine Learning in Medical Image Analysis
Computer-added diagnosis has become an important tool in medicine to support medical doctors in their daily life. The general goals are to classify images to automatically detect diseases or to predict the healing process. Thus, medical imaging builds on several low level tasks such as segmentation, registration, tracking and detection. Many of these tasks can be efficiently solved via machine learning approaches, where, in contrast to typical computer vision problem, we are facing several problems: (1) medical image data such as obtained from CT, MR, or X-ray show specific characteristics (e.g., blur and noise) that cannot easily be handled; (2) machine learning approaches typically require large number of training samples, which is often not available; (3) there are no clear labels as the ground truth is often just based on visual inspection by humans. Thus, there has been a considerable interest in medical image analysis and many approaches have been proposed. As a more comprehensive discussion would be out-of-scope, in the following, we briefly review the most versatile and tools that have been successfully applied in medical image analysis, namely, Support Vector Machines, Random Forests, and Deep Learning.
Support Vector Machines
Support Vector Machines are very versatile tools in machine learning and have thus also be used in medical image analysis for different tasks and applications. In the following, we sketch the main ideas, where we will focus on the two-class classification problem, and give a brief summary of related applications. Let \(\mathbf {\mathcal {L}}=\{(\mathbf {x}_i,y_i)\}_{i=1}^L\) be a set of pairs, where \(\mathbf {x}_i \in \mathbb {R}^N\) are input vectors and \(y_i \in \{+1,-1\}\) their corresponding labels. Then the objective is to determine a linear classification function (i.e., a hyperplane)
where \(\mathbf {w} \in \mathbb {R}^N\), and b is a bias term, such that
which is equivalent to
If the training data is linear separable, then there will exist an infinite number of hyperplanes satisfying Eq. (8). To ensure a unique solution and to increase the linear separability for unseen data (i.e., reduce the generalization error), support vector machines build on the concept of the margin (which is illustrated in Fig. 6), which is defined as the minimal perpendicular distance between a hyperplane and the closest data points. In particular, the decision boundary is chosen such that the margin M is maximized. By taking into account the relation \(\Vert \mathbf {w}\Vert = \frac{1}{M}\), the maximum margin can be obtained my minimizing \(||\mathbf {w}||^2\):
In order to solve the constrained problem Eq. (9) for \(\mathbf w \) and b, we introduce the Lagrange multipliers \(\beta _i, i=1, \dots , L\), and use the Kuhn-Tucker theorem to convert the problem to the unconstrained dual problem (Wolfe dual):
In this way, we get the decision function \(\hat{f}\) for classifying unseen observations \(\mathbf {x}\) as
which is equivalent to
where \(\beta _i > 0\) if \(\mathbf {x}_i\) is on the boundary of the margin, and \(\beta _i = 0\) otherwise. Thus, it can be seen that \(\mathbf {w}\) can be estimated only via a linear combination of samples on the boundary, which are referred to as support vectors (see also Fig. 6).
If the data is not linearly separable, we can apply the kernel trick. As can be seen, Eqs. (10) and (12), the data does only appear in form of dot products \(\langle \mathbf {x}_i, \mathbf {x}_i \rangle = \mathbf {x}_i ^{\top }\mathbf {x}_j\). When introducing a transformation
we need only to estimate the dot product \(\langle \varPhi (\mathbf {x}_i), \varPhi (\mathbf {x}_j) \rangle = \varPhi (\mathbf {x}_i) ^{\top }\varPhi (\mathbf {x}_j)\). Thus, if there is a kernel function
the dot product can be estimated without explicitly knowing \(\varPhi \). Moreover, any other valid kernel can be used, for example:
-
Linear kernel: \(K\left( \mathbf {x}_i, \mathbf {x}_j\right) = \mathbf {x}_i ^{\top }\mathbf {x}_j,\)
-
Polynomial kernel: \(K\left( \mathbf {x}_i, \mathbf {x}_j\right) = \left( \mathbf {x}_i ^{\top }\mathbf {x}_j + 1 \right) ^d,\)
-
Radial Basis Function (RBF) Kernel: \(K\left( \mathbf {x}_i, \mathbf {x}_j\right) = e^{- \gamma \Vert \mathbf {x}_i - \mathbf {x}_j \Vert ^2},\)
-
Mahalanobis kernel: \(K\left( \mathbf {x}_i, \mathbf {x}_j\right) = e^{- (\mathbf {x}_i - \mathbf {x}_j) ^{\top }\mathbf {A} (\mathbf {x}_i - \mathbf {x}_j)}.\)
In this way Eqs. (10) and (12) can be generalized to
and
Besides the flexibility to chose an appropriate kernel for a specific application, it is straightforward to extend the standard formulation for overlapping class distribution by introducing the concept of soft margins. In addition, there exist several ways to extend the standard formulation to multiple classes (e.g., one-vs.-all SVM, pairwise SVM, and error-correcting-output code SVM), to apply SVMs for regression tasks, or to use it in the context of online/incremental and semi-supervised learning. In this way, SVMs are very flexible and widely applicable for the highly diverse task to be solved in medical imaging. For a more detailed review, we like to refer to [37,38,39]).
One of the most important application in medical imaging is to segment and classify image regions. For example, in [40] SVMs are used to segment lesions in ultrasound images. In particular, a kernel SVM using an RBF-kernel is used to segment both ultrasound B-mode and clinical ultrasonic images. Similarly, in [41] an effective retinal vessel segmentation technique is presented, allowing to drastically reduce the manual effort of ophthalmologists. To this end, first features are extracted which are then classified using a linear SVM. This makes not only the evaluation very fast, but also allows to learn a model form a smaller training set.
A different approach is followed in [42] to segment blood vessels based on fully connected conditional random fields. However, an efficient inference approach is applied, which is learned via a Structured Output SVM. In this way, a fully automated system is obtained that achieves human-like results. Similarly, [43] presents a fully automatic method for brain tissue segmentation, where the goal is to segment 3D MRI images of brain tumor patients into healthy and tumor areas, including their individual sub-regions. To this end, an SVM classification using multi-spectral intensities and textures is combined with a CRF regularization.
A slightly different application in medical imaging is to localize image regions. For example, [44] presents an approach to detect microcalcification (MC) clusters in digital mammograms via an SVM-based approach. This is in particular of interest, as MC clusters can be an early indicator for female breast cancer. A different application, but a similar approach was discussed in [45]. The goal is to localize the precise location of cell nuclei, helping in an automated microscopy applications such as such as cell counting and tissue architecture analysis. For this purpose three different inputs are used (i.e., raw pixel values, edge values, and the combination of both), which are used to train an SVM classifier based on an RBF-kernel.
Random Forests
Random Forests (RFs) [46], in general, are ensembles of decision trees, which are independently trained using randomly drawing samples from the original training data. In this way, they are fast, easy to parallelize, and robust to noisy training data. In addition, they are very flexible, paving the way for classification, regression, and clustering tasks, thus making them a valid choice for a wide range of medical image applications [47].
More formally, Random Forests are ensembles of T decision trees \(\mathcal {T}_t(\mathbf {x}): \mathcal {X} \rightarrow \mathcal {Y}\), where where \(\mathcal {X} = \mathbb {R}^N\) is the N-dimensional feature space and \(\mathcal {Y}\) is the label space \(\mathcal {Y} = \{1, \dots , C\}\). A decision tree can be considered a directed acyclic graph with two different kinds of nodes: internal (split) nodes and terminal (leaf) nodes. Provided a sample \(\mathbf {x} \in \mathcal {X}\), starting from the root node at each split node a decision is made to which child node the sample should be send, until it reaches a leave node. Each leaf note is associated with a model that assigns an input \(\mathbf {x}\) an output \(y \in \mathcal {Y}\). Each decision tree thus returns a class probability \(p_t(y|\mathbf {x})\) for a given test sample \(\mathbf {x} \in \mathbb {R}^N\), which is illustrated in Fig. 7(b). These probabilities are then averaged to form the final class probabilities of the RF. A class decision for a sample \(\mathbf x \) is finally estimated by

During training of a RF, each decision tree is provided with a random subset of the training data \(\mathcal {D} = \left\{ (x_1, y_1), \dots (x_{|D|}, y_{|D|}) \right\} \subseteq \mathcal {X} \times \mathcal {Y}\) (i.e., bagging) and is trained independently from each other. The data set \(\mathcal {D}\) is then recursively split in each node, such that the training data in the newly created child nodes is pure according to the class labels. Each tree is grown until some stopping criterion (e.g., a maximum tree depth) is met and class probability distributions are estimated in the leaf nodes. This is illustrated in Fig. 7(a).
Random Forests: (a) The tree is build recursively splitting the training data \(\mathcal {D}\) and finally estimating a model \(p(y|\mathcal {D}_{*})\) for each leaf node. (b) During inference a sample \(\mathbf x \) is traversed down according to the learned splitting functions s (i.e., the parameters \(\varTheta ^{*}\)) the tree and finally classified based on the model of the leaf node.
A splitting function \(s(\mathbf {x}, \varTheta )\) is typically parameterized by two values: (i) a feature dimension \(\varTheta _1 \in \{1, \ldots , N\}\) and (ii) a threshold \(\varTheta _2 \in \mathbb {R}\). The splitting function is then defined as
where the outcome defines to which child node the sample \(\mathbf {x}\) is routed.
Each node i chooses the best splitting function \(\varTheta ^i\) out of a randomly sampled set by optimizing the information gain
where \(\mathcal {D}_L\) and \(\mathcal {D}_R\) are the sets of data samples that are routed to the left and right child nodes, according to \(s(\mathbf {x}, \varTheta ^i)\); \(H(\mathcal {D})\) is the local score of a set \(\mathcal {D}\) of data samples, which can either be the negative entropy
where C is the number of classes, and p(c|S) is the probability for class c, estimated from the set S, or the Gini Index [46].
The most important application of RFs in medical image analysis is the automatic segmentation of cells, organs or tumors, typically building on a multi-class classification forests. For example, in [48] an approach for segmenting high-grade gliomas and their sub-regions from multi-channel MR images is presented. By using context-aware features and the integration of a generative model of tissue appearance only little pre-processing and no explicit regularization is required, making the approach computationally very efficient. A different approach was presented in [49], where a joint classification-regression forest was trained, that captures both structural and class information. In this way, not only a class label is predicted but also the distance to the object boundary. Applied on 3-dimensional CT scans the final task of multi-organ segmentation can be solved very efficiently.
Related to the previous task is the application of detecting and localizing anatomy. For example, [50] introduces an approach for localizing vertebras using a combined segmentation and localization approach. For this purpose a RF is trained using features images obtained form a standard filter bank, where the output is then used – together with the original image – to generate candidate segmentations for each class, which are finally weighted. In contrast, [51] addresses the problem of localizing organs such as spleen, liver or heart. To this end, visual features are extracted from the imaging data and a regression forest is trained, allowing for a direct mapping form voxels to organ locations and size. In particular, the approach deals with both magnetic resonance (MR) and computer tomography (CT) images, also showing the generality and flexibility of RFs. A similar approach is addressed in [52], also estimating local landmark points finally paving the way for automatic age estimation [53].
For a detailed overview on Random Forests we would like to refer to [47], where a deep theoretical discussion as well as an overview of different applications in the field of medical image analysis are given.
Deep Learning
Event though the main ideas of neural networks are dating back to the 1940’s (i.e., [54, 55]), they just become recently very popular due the success of convolutional neural networks [56, 57]. In general, neural networks, are biologically inspired and can be described as a directed graph, where the nodes are related to neurons/units and the edges describe the links between them.
As illustrated in Fig. 8, each unit j receives a weighted sum of inputs \(a_i\) of connected units i, where the weights \(w_{i,j}\) determine the importance of the connection. To estimate the output \(a_j\) this linear combination is then fed into a so called activation function. More formally, the output \(a_j\) is estimated as follows:
Popular choices for the activation function which are widely used are
-
Linear function: \(g(x) = x\),
-
Hard threshold function: \(g(x) = \left\{ \begin{array}{ll} 1 &{} \quad \text {if} \; x > \frac{1}{2} \\ 0 &{} \quad \text {otherwise}, \end{array}\right. \)
-
Sigmoid function: \(g(x) = \frac{1}{1+e^{-x}}\),
-
Hyperbolic tangent function: \(g(x) = tanh(x)\),
-
Rectified Linear Units (ReLU): \(g(x) = max(0,x)\).
In general, we can distinct two different kinds of networks. First, feed-forward networks can be described as acyclic graphs, having connections only in one direction. In this way, the network describes a function of the input. In contrast, recurrent networks (RNNs) can be considered graphs with loops, as receiving their outputs again as input (thus being non-linear systems). In this way, an internal state (short term memory) can be described. Thus, RNNs are widely used in applications such as speech recognition or in activity recognition [58], whereas in image processing mainly feed-forward networks are of relevance [59].
Neural networks are typically arranged in layers \(V_i\) consisting of single units as described above, such that each unit receives input only from units from the previous layer, where \(|V| = T\) the depth of the network. \(V_0\) is referred to as the input layer, \(V_T\) as the output layer, and \(V_1, \dots , V_{T-1}\) are called the hidden layers. A simple example of such a network with two hidden layers is illustrated in Fig. 9. When dealing with multiple hidden layers, we talk about deep learning. In general, this allows to learn complex functions, where different layers cover different kind of information. For example, in object detection a first layer may describe oriented gradients, a second layer some kind of edges, a third layer would assemble those edges to object descriptions, where a subsequent layer would describe the actual detection task. This example also illustrates an important property of deep learning: we can learn feature representations and do not need to design features by hand!
In general, the goal of supervised learning is to modify the model parameters such that subsequently the error of an objective function is reduced. For neural networks, this is typically solved via the stochastic gradient descend (SGD) approach. The main idea is to repeatedly compute the errors for many small sets and to adjust the model according to a averaged response. Thus, the name can be explained as a gradient method – typically using the back-propagation approach – is used and the computation based on small sets of samples is naturally noisy.
The most prominent and most successful deep learning architecture are Convolutional Neural Networks (CNN), why these terms are often used interchangeable. Even though naturally inspired by image processing problems the same ideas can also be beneficial for other tasks. One key aspect of CNNs is that the are structured in a series of different layers: convolutional layers, pooling layers, and fully connected layers. Convolutional layers can be considered feature maps, where each feature map is connected to local patches in the feature map in the previous layer. In contrast, pooling layers merge similar features into one (i.e., relative positions of features might vary in the local range). Typical, several stages of convolutional and pooling layers are stacked together. Finally, there are fully connected layers generating the output of the actual task. A typical architecture for such a CNN is illustrated in Fig. 10.
Typical convolutional neural network: LeNet-5 [56].
As in this way the effort for handcrafting features can be reduced and CNNs have proven to improve the results for many applications, they are now also widely applied in medical imaging. In particular, for cancer detection very recently even human-like performance was demonstrated.
For example, [60] adopts a CNN framework for breast cancer metastasis detection in lymph nodes. By additionally, exploiting the information of a pre-trained model, sophisticated image normalization, and building on a multi-stage approach (mimicking the human perception), state-of-the-art methods and even human pathologists have been outperformed for a standard benchmark dataset. Similarly, [61] addresses the problem of skin cancer detection using deep neural networks. Also here a transfer learning setup is proposes, where after pre-training a CNN architecture using ImageNet the final classification layer is discarded and re-trained for the given task (in addition the parameters are fine-tuned across all layers). The thus obtained automatic methods finally performs on par with human pathologist on different task.
Even though this demonstrates, that Deep Learning could be very beneficial in the medical domain, the main challenge is to cope with the problem that often the rather large amount of required training data is not available. Thus, there has been a considerable interest in approaches that can learn from a small number of training samples. The most common and straight forward way is to use data augmentation, where additional training samples are generated via variation of the given data: rotation, elastic deformation, adding noise, etc. One prominent example for such approaches is U-Net [62], which demonstrated that for biomedical image segmentation state-of-the-art results can be obtained, even when the model was trained just from a few samples.
Even though this simple approach often yields good results, it is limited as only limited variations can be generated from the given data. A different direction is thus to build on ideas from transfer learning [63]. The key idea is to pre-train a network on large publicly available datasets and then to fine-tune it for the given task. For example, [64] fine-tunes the VGG-16 network, which is already pre-trained using a huge amount of natural images, to finally segment pancreas from MR images. In addition, a CRF step is added for the final segmentation. Another way would be to use specific prior knowledge about the actual task [65]. However, this information is often not available and, as mentioned above, medical image data and natural images are often not sharing the same characteristics, why such approaches often to fail in practice.
A totally different way to deal with small amounts of training data is to use synthetically generated samples for training (e.g., [66]), which are easy to obtain. However, again in this way the specific characteristic of the given image data might not be reflected. To overcome this problem, Generative Adversarial Nets [67] train a generator and a discriminator framework in a competitive Random Forests fashion. The key idea is that the generator synthesizes images and the discriminator decides if an image is real or fake (i.e., generated by the generator). In this way, increasingly better training data can be generated. This idea is for example exploited by [68] to better model the nonlinear relationship between CR and MR images.
Further Reading: For a short review on Deep Learning we would like to refer to [13], a detailed review of related work can be found in [69], and a very detailed technical overview ins given in [70].
Summary
As demonstrated in this section there are several ways to address the wide range of applications in medical imaging. Even though there exist special approaches for specific application, we focused on three versatile and thus widely used approaches, namely, Support Vector Machines (SVMs), Random Forests (RFs), and Deep Learning (DL). Where SVMs are general working horses for different applications, RFs have demonstrated to cope with the particular characteristics of medical imaging data very well. However, for both approaches well-engineered handcrafted features are necessary, which are often hard to define and compute. This problem can be overcome by using DL approaches, as the required features can be learned implicitly in an end-to-end manner. However, the main drawback of such approaches is that they require a huge amount of training data to yield competitive results, which is often not available in practice. There are several approaches which help to moderate this problem, but in general dealing with a small data is still a big problem. Thus, still other methods such as SVM and RF are valid choices for medical imaging problems. In addition, a key aspect that is often neglected is that there are often good biases available, either defined by the specific task or by available human experts, which are not considered (sufficiently) up to now!
5 Secure Cooperation with Experts
Securing the data life cycle is a problem that just recently gained a lot of additional attention, mainly due to the General Data Protection Regulation (GDPR)Footnote 1 that not only established baselines for securing sensitive information throughout the European Union, but also increases the penalties to be applied in case of violation. This regulation will come into effect in May 2018 either directly or by adoption into national law. It is concerned with all major issues regarding the processing of personal sensitive information, most notably it deals with the legal requirements regarding data collection, consent regarding processing, anonymization/pseudonymization, data storage, transparency and deletion [71]. Still, the major issue here is that many details are currently not defined, e.g. whether deletion needs to be done on a physical or simply logical level, or how strong the anonymization-factors need to be [72]. Furthermore, some parts are formulated in a way that cannot be achieved with current technological means, e.g. de-anonymization being impossible in any case, as well as the antagonism between deletion and transparency. Thus, the issue of securing sensitive information is one of the really big challenges in machine learning in health related environments [73].
5.1 Data Leak Detection
While many of the issues outlined above seem not that relevant for pathological data at first glance, other questions regarding data security still prevail: Data is considered to be the new oil, meaning that data itself constitutes a significant value. One of the major issues in machine learning based research lies in the issue of cooperation between data owners and other entities. With ever new techniques arriving on the scene requiring the cooperation of various experts in the areas of modeling, machine learning and medicine, data needs to be shared between different entities, often working at different institutions. This opens up the issue of detecting data misuse, especially the unsolicited dissemination of data sets.
Measures against data leakage can be divided into two major categories, those protecting the data from being leaked (proactive measures) and those enabling the detection and attribution of data leaks (reactive measures). Proactive measures typically include limitations on the data exchange:
-
Sealed research environments like dedicated servers that run all the analysis software and contain all the data, without export possibilities, as well as sporting mechanisms for controlling, which researcher utilized which information set. While this does not constitute a 100 percent protection against malicious experts trying to extract information, in does help against accidental data loss.
-
Aggregating the data as much as possible can be a solution too for reducing the amount of sensitive information that an expert is given access to. Still, aggregation often introduces a significant error and can render the data practically worthless for the analysis.
-
Oracle-style measures like differential privacy [74] do not deliver the whole data set to the experts but rather require the expert to choose the type of analysis he/she wants to run on the data without seeing the data sets. Typically, this is done via issuing “Select”-statements that are run against the database. Measures like differential privacy introduce distortion for data protection purposes, as well as limit the amount of information the expert can retrieve from the data.
While these proactive measures certainly do have their merits, they often pose a serious obstacle to cooperation with the world-best experts in a field, either due to geographical issues, or simply because the data is not fine-grained enough to utilize the whole potential of the information inside the data sets. Reactive approaches aim at identifying data leaks instead of preventing exchange, i.e. the data is distributed to the partners in a form suitable for analysis (while, of course, still considering issues of data protection like required anonymization), but it is marked in order to make each data set unique for each partner it is distributed to. These marks are typically called fingerprints and are required to possess the following features [75]:
-
The fingerprinting mechanism must be capable to uniquely identify a user.
-
It must not be possible for a user to identify the mark and subsequently remove it (without drastically reducing the utility of the data).
-
Even for a combination of attackers, the fingerprint must be resilient against inference attacks, i.e. it must not be possible to calculate the fingerprinting marks, even when given several differently marked versions of the same data set.
-
No wrongful accusations must be possible, even in case several attacks work together in order to change the fingerprint to put blame on an uninvolved partner.
-
The fingerprint must be tolerant against a certain amount of errors introduced by the users, i.e. it must not get useless in case the attackers change some portion of the marked data.
Furthermore, depending on the actual form of the data, there are typically some additional issues that require consideration, e.g., how stable the fingerprint needs to be in case only part of the data is leaked (e.g. half a picture, some records from a sample). Since the GDPR often requires the anonymization of sensitive information, one interesting approach lies in the development of combined methods that use intrinsic features of the anonymization technique in order to generate a selection of different data sets that can be uniquely identified. In the past, such a fingerprinting approach was proposed for structured data in tables [76], still, the number of different fingerprints that can be assigned to the same data set while providing resilience against collaborating attackers is rather low and mainly depends on the actual data, especially when obeying the requirement for resilience against colluding attackers as outlined above [77].
5.2 Deletion of Data
Typically, the deletion of data is not considered to be a major problem in most applications, as it is mostly a matter of freeing resources. Still, against the background of the GDPR, this topic becomes increasingly important to consider, especially, since it is extremely complicated to delete information in modern, complex environments [72]. Databases are a major example, why deletion of information can be a major problem: ACID-compliance [78] is a major requirement of modern database management systems and requires the database product to ensure the atomicity of certain operations, i.e. operations are either carried out as a whole, or not at all, always leaving the database to be in a consistent state, even in case of a server crash. Furthermore, mechanisms for undoing operations, most notable rollback mechanisms, are currently state-of-the-art and expected by database users. Thus, a lot of additional information is required to be stored in various internal mechanisms of the database, e.g. the transaction mechanism, which is responsible for collecting all operations changing the database and enables rollbacks and crash-recovery.
Still, in typical database environments, even simple removal of data from the database itself is non-trivial, considering the way data is stored: The records inside a table are stored inside a tree-structure, more notably a \(B^+\)-tree [79]:
-
For the number \(m_i\) of elements of node i holds \(\frac{d}{2}\le i \le d\), given a pre-defined d for the whole tree, the order of the tree. The ony example of this rule is the root r with \(0\le r \le d\).
-
Each non-leaf-node with m elements possesses \(m+1\) child nodes, \(\frac{d}{2}\le m\le d\).
-
The tree is balanced, i.e. all leaf nodes are on the same level.
-
In contrast to the normal B-tree, the inner nodes of the \(B^+\)-tree solely store information required for navigating through the tree, the actual data is stored in the leaf nodes, making the set of leafs forming a partition of the whole data set.
-
The elements inside the nodes are stored as sorted lists.
In databases like MySQL, the (mandatory) primary key of each table is used to physically organize the data inside the table in the form of a \(B^+\)-Tree, the secondary indices are merely search-trees of their own, solely containing links to the tree built by the primary key.
When data is deleted from the indexing tree built by the primary key, the database searches for the leaf node containing the required element. Since databases are built in order to facilitate fast operations, the data inside the leaf node is not overwritten, but simple unlinked from the sorted list inside said node.
Deletion in MySQL [80].
Figure 11 gives a short overview on the deletion process: The record in question is unlinked from the linked list and added to the so-called garbage collection, which marks the space of the record as free for storing new data. Still, the data is not technically overwritten at the point of deletion and can be reconstructed quite simple, as long as the space has not been used again, which depending on the usage patterns of the database, is unpredictable and might take a long time.
Still, even the actual form of the search tree itself might yield information on data already deleted from the table [81]: Let B be a \(B^+\)-tree with \(n>b\) elements which are added in ascending order. Then it holds true that the partition of the leafs of B has the following structure:
While this theorem allows only very limited detection for practical purposes under certain circumstances, especially due to database internal reorganization mechanisms destroying the structure, there are instances where information can be retrieved from the structure of the \(B^+\)-tree.
As [72] outlined, there is currently no definition in the GDPR, how data must be deleted, i.e. whether this needs to be done physically, or only logically. Still, when looking at earlier national legal counterparts concerning data protection, several legislations (e.g. in Austria) used a very strict interpretation of the term “deletion”, i.e. physical deletion. In addition, the GDPR deals a lot in absolutes, either jurisdiction is required to relax these absolutes, or new technical means for removing evidence deeply embedded in complex systems are required.
5.3 Information Extraction
We have already seen how databases can be secured via differential privacy and other query mechanisms, however most data in a clinical environment exist in the form of Electronic Health records whose entries are mostly unstructured free-text documents. As there is no guaranteed way to anonymize unstructured data, we first need to extract identified bits of information and convey them to a more organized data structure.
Information Extraction (IE) is the art of finding relevant bits of specific meaning within unstructured data - this can be done either via (1) low-level IE - usually by means of dictionary or RegExp based approaches [82, 83] which utilize extensive corpora of biomedical vocabulary and are readily available in libraries such as Apache cTakes; the disadvantage of most standard solutions is the lack of their ability to correctly identify context, ambiguous, synonymous, polysemous or even just compound words; or (2) higher level IE, usually in the form of a custom-built natural language processing (NLP) pipelines. Often, the purpose of such pipelines is Named Entity Recognition (NER), which is the task of labeling terms of specific classes of interest, like People, Locations, or Organizations.
It was noted [84] that NER is more difficult in specialized fields, as terms have more narrow meanings (abbreviations can mean different things, e.g.). The authors of [85] describe NER as a sequence segmentation problem to which they apply Conditional Random Fields (CRF), a form of undirected statistical graphical models with Markov independence assumption, allowing them to extract orthographic as well as semantic features. More recently, even Neural Networks have been utilized for NER [86] with performance at state-of-the-art levels, partly incorporating a new form of concept space representations for terms called embeddings, which use a form of dimensionality reduction to compress vocabulary-sized feature vectors into (mostly 50-300 dimensional) concept vectors [87].
5.4 Anonymization
After having condensed all available material into labeled information, we can filter them through formal anonymization approaches, of which k-anonymity [88] stands as the most prominent. K-anonymity requires that a release of data shall be clustered into equivalence groups of size \(>=k\) in which all quasi identifiers (non-directly identifying attributes such as age, race or ZIP code) have been generalized into duplicates; generalization itself can be pictured as an abstraction of some information to a more inclusive form, e.g. abstracting a ZIP code of 81447 to textit81***, thereby being able to potentially cluster it with all other ZIP codes starting with 81***.
Beyond k-anonymity exist refinements such as l-diversity [89], t-closeness [90] and \(\delta \)-presence [91] for purely tabular data, as well as a number of individual methods for social network anonymization [92, 93]. They all operate on the concept of structured, textual quasi identifiers and imply a trade-off between data utility and privacy of a data release - a higher degree of anonymization provides more security against identifying a person contained in the dataset, but reduces the amount of usable information for further studies or public statistics.
This leads us to the field of Privacy aware Machine Learning (PaML) which is the application of ML techniques to anonymized (or in any way perturbed) datasets. Obviously one cannot expect the performance of such algorithms to equal their counterparts executed on the original data [94], instead the challenge is to produce anonymized datasets which yield results of similar quality than the original. This can be achieved by cleverly exploiting statistical properties of such data, e.g. outliers in the original might affect ML performance as well as induce higher levels of generalization necessary to achieve a certain factor of k; by first removing those outliers an anonymized version can actually retain enough information to rival its unperturbed predecessor [95].
5.5 Image Data Integration
For medical purposes, images have long been considered quasi-identifiers [96, 97], as one can easily picture faces allowing a relatively exact reconstruction of a persons identity (depending on the quality of algorithms used). In the case of pathological images containing multiple features and feature groups in relation to one another, any subset of such information could conceivably be combined with a patient’s EHR, thus enabling re-identification. On the other hand, selected features also complement a patients EHR and therefore provide a more complete overview of the patient’s condition facilitating higher precision in diagnosis, especially in cases when doctors overlook or forget to record symptoms. In approximating an answer to the question’how can one anonymize images’, we would like to provide a simple (therefore unrealistic) illustration (Fig. 12), in which the bottom row depicts 8 original face images, whereas the subsequent vertical rows represent progressive morphings of pairs of samples below, arriving at a most general male/female hybrid at the top. In a realistic clinical setting, a useful effect could be achieved by learning features from individual samples (faces, pathologic slides etc.), clustering those traits by similarity and then merging them together into collective representations of groups.
Depiction of a possible (naive) face generalization hierarchy by simple morphing of aligned images. Realistically, one would first scan images for salient features, then cluster faces via feature similarity, subsequently morphing the generalized features back into a pictograph or artificially generated face.
5.6 *Omics Data Integration
In contrast to images it seems very doubtful if *omics-data can be perturbed in a meaningful way to protect a person’s identity. After all, genes, proteins etc. are building blocks of larger structures, and changing even one gene to a variant form (called an allele) can have significant repercussions on an organism’s phenotype. So in the field of *omics research, the issue of privacy is treated a little differently: Given e.g. a GWAS (genome-wide association study) and the genetic profile of an individual person, the question arises with what certainty a classifier could determine if that person participated in said study. This entails the need to perturb the results of a study - a distribution of measurements, like allele frequencies [98] - rather than a database of personal information, which lends itself ideally to the already described mechanism of \(\epsilon \)-differential privacy. The authors of [99] even tailored the method to GWAS study data in case of the presence of population stratification and studied its effect on the output of the EIGENSTRAT and LMM (Linear Mixed Model) algorithms typically used on a rheumatoid arthritis GWAS dataset. To what extent those methods can actually protect people from identification is a point of open discussion: while some researchers [100] claim that even with standard statistical methods a binary classification result (AUC) of reasonably close to 1 can be achieved, others [101] point out that DNA matching in itself is not equivalent to de-identification and even if possible, would take tremendous time and computational power. It might therefore be the case that a false notion of a choice of privacy OR data utility might lead to a gross over-expansion of the privacy legal framework.
5.7 Heterogeneous Data Linkage
As became obvious from the previous sections, information about the same person are often available from several different sources (physician letters, hospital databases, lab reports, scans, *omics data etc.). These data are not easily merged into one big dataset because coverage might only be slightly overlapping (e.g. not all patients were subjected to the same lab tests). Simple concatenation of such information would result in a high-dimensional dataset with most of its entries missing, introducing the curse-of-dimensionality when conducting ML experiments. With increasing dimensionality, the volume of the space increases so fast that the available data becomes sparse, hence it becomes impossible to find reliable clusters; also the concept of distance becomes less precise as the number of dimensions grows, since the distance between any two points in a given data set converges; moreover, different clusters might be found in different sub spaces, so a global filtering of attributes is also not sufficient. A solution might be found in graph-based representations of such data, where node types can represent patients or different forms of examinations, resources, etc.; in the case of anonymizing, we not only have to generalize node information but also consider neighborhood structure which could provide an adversary with additional hints for attack vectors. Apart from dealing with graph anonymization, which is also a hard problem [102], an interesting challenge lies in describing parameters of the underlying stochastic process precisely enough so one can re-populate a graph from its anonymized form; this generatively perturbed graph should on the one hand meet privacy requirements, yet allow scientists to conduct ML experiments yielding satisfactory performance.
6 Future Challenges
Much future research has to be done, particularly in the fields of Multi-Task Learning and Transfer Learning to go towards Multi-Agent-Hybrid Systems as applications of the iML-approach.
6.1 Future Challenge 1: Multi-task Learning
Multi-task learning (MTL) aims to improve the prediction performance by learning a problem together with multiple, different but related other problems through shared parameters or a shared representation. The underlying principle is bias learning based on Probably Approximately Correct learning (PAC learning) [19]. To find such a bias is still the hardest problem in any ML task and essential for the initial choice of an appropriate hypothesis space, which must be large enough to contain a solution, and small enough to ensure a good generalization from a small number of data sets. Existing methods of bias generally require the input of a human-expert-in-the-loop in the form of heuristics and domain knowledge to ensure the selection of an appropriate set of features, as such features are key to learning and understanding. However, such methods are limited by the accuracy and reliability of the expert’s knowledge (robustness of the human) and also by the extent to which that knowledge can be transferred to new tasks (see next subsection). Baxter (2000) [103] introduced a model of bias learning which builds on the PAC learning model which concludes that learning multiple related tasks reduces the sampling burden required for good generalization. A bias which is learnt on sufficiently many training tasks is likely to be good for learning novel tasks drawn from the same environment (the problem of transfer learning to new environments is discussed in the next subsection). A practical example is regularized MTL [104], which is based on the minimization of regularization functionals similar to Support Vector Machines (SVMs), that have been successfully used in the past for singletask learning. The regularized MTL approach allows to model the relation between tasks in terms of a novel kernel function that uses a taskcoupling parameter and largely outperforms singletask learning using SVMs. However, multi-task SVMs are inherently restricted by the fact that SVMs require each class to be addressed explicitly with its own weight vector. In a multi-task setting this requires the different learning tasks to share the same set of classes. An alternative formulation for MTL is an extension of the large margin nearest neighbor algorithm (LMNN) [105]. Instead of relying on separating hyper-planes, its decision function is based on the nearest neighbor rule which inherently extends to many classes and becomes a natural fit for MTL. This approach outperforms state-of-the-art MTL classifiers, however, much open research challenges remain open in this area [106].
6.2 Future Challenge 2: Transfer Learning
A huge problem in ML is the phenomenon of catastrophic forgetting, i.e. when a ML algorithm completely and abruptly “forgets” how to perform a learned task once transferred to a different task. This is a well-known problem which affects ML-systems and was first described in the context of connectionist networks [107]; whereas natural cognitive systems rarely completely disrupt or erase previously learned information, i.e. natural cognitive systems do not forget “catastrophically” [108]. Consequently the challenge is to discover how to avoid the problem of catastrophic forgetting, which is a current hot topic [109].
According to Pan and Yang (2010) [21] a major requirement for many ML algorithms is that both the training data and future (unknown) data must be in the same feature space and show similar distribution. In many real-world applications, particularly in the health domain, this is not the case: Sometimes we have a classification task in one domain of interest, but we only have sufficient training data in another domain of interest, where the latter data may be in a completely different feature space or follows a different data distribution. In such cases transfer learning would greatly improve the performance of learning by avoiding much expensive data-labeling efforts, however, much open questions remain for future research [110].
6.3 Future Challenge 3: Multi-Agent-Hybrid Systems
Multi-Agent-Systems (MAS) are collections of many agents interacting with each other. They can either share a common goal (for example an ant colony, bird flock, or fish swarm etc.), or they can pursue their own interests (for example as in an open-market economy). MAS can be traditionally characterized by the facts that (a) each agent has incomplete information and/or capabilities for solving a problem, (b) agents are autonomous, so there is no global system control; (c) data is decentralized; and (d) computation is asynchronous [111]. For the health domain of particular interest is the consensus problem, which formed the foundation for distributed computing [112]. The roots are in the study of (human) experts in group consensus problems: Consider a group of humans who must act together as a team and each individual has a subjective probability distribution for the unknown value of some parameter; a model which describes how the group reaches agreement by pooling their individual opinions was described by DeGroot [113] and was used decades later for the aggregation of information with uncertainty obtained from multiple sensors [114] and medical experts [115]. On this basis Olfati-Saber et al. [116] presented a theoretical framework for analysis of consensus algorithms for networked multi-agent systems with fixed or dynamic topology and directed information flow. In complex real-world problems, e.g. for the epidemiological and ecological analysis of infectious diseases, standard models based on differential equations very rapidly become unmanageable due to too many parameters, and here MAS can also be very helpful [117]. Moreover, collaborative multi-agent reinforcement learning has a lot of research potential for machine learning [118].
7 Conclusion
Machine learning for digital pathology poses a lot of challenges, but the premises are great. An autonomous pathologist, acting as digital companion to augment real pathologists can enable disruptive changes in future pathology and in whole medicine. To reach such a goal much further research is necessary in collecting, transforming and curating explicit knowledge, e.g. clinical data, molecular data and e.g. lifestyle information used in medical decision-making.
Digital Pathology will highly benefit from interactive Machine Learning (iML) with a pathologist in the loop. Currently, modern deep learning models are often considered to be “black-boxes” lacking explicit declarative knowledge representation. Even if we understand the mathematical theories behind the machine model it is still complicated to get insight into the internal working of that model, hence black box models are lacking transparency and the immediate question arises: “Can we trust our results?” In fact: “Can we explain how and why a result was achieved?” A classic example is the question “Which objects are similar?”, but an even more interesting question is “Why are those objects similar?”. Consequently, in the future there will be urgent demand in machine learning approaches, which are not only well performing, but transparent, interpretable and trustworthy. If human intelligence is complemented by machine learning and at least in some cases even overruled, humans must be able to understand, and most of all to be able to interactively influence the machine decision process. A huge motivation for this approach are rising legal and privacy aspects, e.g. with the new European General Data Protection Regulation (GDPR and ISO/IEC 27001) entering into force on May, 25, 2018, will make black-box approaches difficult to use in business, because they are not able to explain why a decision has been made.
This will stimulate research in this area with the goal of making decisions interpretable, comprehensible and reproducible. On the example of digital pathology this is not only useful for machine learning research, and for clinical decision making, but at the same time a big asset for the training of medical students. Explainability will become immensely important in the future.
Notes
- 1.
Regulation (EU) 2016/679 of the European Parliament and of the council of 27 April 2016 on the protection of natural persons with regard to the processing of personal data and on the free movement of such data, and repealing directive 95/46/EC (General Data Protection Regulation).
References
Madabhushi, A., Lee, G.: Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 33, 170–175 (2016)
Holzinger, A.: Trends in interactive knowledge discovery for personalized medicine: Cognitive science meets machine learning. IEEE Intell. Inf. Bull. 15, 6–14 (2014)
Su, X., Kang, J., Fan, J., Levine, R.A., Yan, X.: Facilitating score and causal inference trees for large observational studies. J. Mach. Learn. Res. 13, 2955–2994 (2012)
Huppertz, B., Holzinger, A.: Biobanks – a source of large biological data sets: open problems and future challenges. In: Holzinger, A., Jurisica, I. (eds.) Interactive Knowledge Discovery and Data Mining in Biomedical Informatics. LNCS, vol. 8401, pp. 317–330. Springer, Heidelberg (2014). doi:10.1007/978-3-662-43968-5_18
Mattmann, C.A.: Computing: A vision for data science. Nature 493, 473–475 (2013)
Otasek, D., Pastrello, C., Holzinger, A., Jurisica, I.: Visual data mining: effective exploration of the biological universe. In: Holzinger, A., Jurisica, I. (eds.) Interactive Knowledge Discovery and Data Mining in Biomedical Informatics. LNCS, vol. 8401, pp. 19–33. Springer, Heidelberg (2014). doi:10.1007/978-3-662-43968-5_2
Ghahramani, Z.: Probabilistic machine learning and artificial intelligence. Nature 521, 452–459 (2015)
Teh, Y.W., Jordan, M.I., Beal, M.J., Blei, D.M.: Hierarchical dirichlet processes. J. Am. Stat. Assoc. 101, 1566–1581 (2006)
Houlsby, N., Huszar, F., Ghahramani, Z., Hernndez-lobato, J.M.: Collaborative gaussian processes for preference learning. In: Pereira, F., Burges, C., Bottou, L., Weinberger, K. (eds.) Advances in Neural Information Processing Systems (NIPS 2012), pp. 2096–2104 (2012)
Holzinger, A.: Introduction to machine learning and knowledge extraction (make). Mach. Learn. Knowl. Extr. 1, 1–20 (2017)
Shahriari, B., Swersky, K., Wang, Z., Adams, R.P., de Freitas, N.: Taking the human out of the loop: A review of bayesian optimization. Proc. IEEE 104, 148–175 (2016)
Kim, W., Choi, B.J., Hong, E.K., Kim, S.K., Lee, D.: A taxonomy of dirty data. Data Min. Knowl. Disc. 7, 81–99 (2003)
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521, 436–444 (2015)
Lee, S., Holzinger, A.: Knowledge discovery from complex high dimensional data. In: Michaelis, S., Piatkowski, N., Stolpe, M. (eds.) Solving Large Scale Learning Tasks. Challenges and Algorithms. LNCS (LNAI), vol. 9580, pp. 148–167. Springer, Cham (2016). doi:10.1007/978-3-319-41706-6_7
Holzinger, A., Plass, M., Holzinger, K., Crisan, G.C., Pintea, C.M., Palade, V.: A glass-box interactive machine learning approach for solving NP-hard problems with the human-in-the-loop (2017). arXiv:1708.01104
Holzinger, A.: Interactive machine learning for health informatics: When do we need the human-in-the-loop? Brain Informatics (BRIN) 3 (2016)
Holzinger, A., Plass, M., Holzinger, K., Crişan, G.C., Pintea, C.-M., Palade, V.: Towards interactive machine learning (iML): applying ant colony algorithms to solve the traveling salesman problem with the human-in-the-loop approach. In: Buccafurri, F., Holzinger, A., Kieseberg, P., Tjoa, A.M., Weippl, E. (eds.) CD-ARES 2016. LNCS, vol. 9817, pp. 81–95. Springer, Cham (2016). doi:10.1007/978-3-319-45507-5_6
Fayyad, U., Piatetsky-Shapiro, G., Smyth, P.: From data mining to knowledge discovery in databases. AI Mag. 17, 37–54 (1996)
Valiant, L.G.: A theory of the learnable. Commun. ACM 27, 1134–1142 (1984)
Holzinger, A.: On topological data mining. In: Holzinger, A., Jurisica, I. (eds.) Interactive Knowledge Discovery and Data Mining in Biomedical Informatics. LNCS, vol. 8401, pp. 331–356. Springer, Heidelberg (2014). doi:10.1007/978-3-662-43968-5_19
Pan, S.J., Yang, Q.: A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2010)
Demichelis, F., Barbareschi, M., Dalla Palma, P., Forti, S.: The virtual case: a new method to completely digitize cytological and histological slides. Virchows Arch. 441, 159–161 (2002)
Bloice, M., Simonic, K.M., Holzinger, A.: On the usage of health records for the design of virtual patients: a systematic review. BMC Med. Inform. Decis. Mak. 13, 103 (2013)
Turkay, C., Jeanquartier, F., Holzinger, A., Hauser, H.: On computationally-enhanced visual analysis of heterogeneous data and its application in biomedical informatics. In: Holzinger, A., Jurisica, I. (eds.) Interactive Knowledge Discovery and Data Mining in Biomedical Informatics. LNCS, vol. 8401, pp. 117–140. Springer, Heidelberg (2014). doi:10.1007/978-3-662-43968-5_7
Ferreira, R., Moon, B., Humphries, J., Sussman, A., Saltz, J., Miller, R., Demarzo, A.: The virtual microscope. In: Proceedings of the AMIA Annual Fall Symposium, pp. 449–453 (1997)
Barbareschi, M., Demichelis, F., Forti, S., Palma, P.D.: Digital pathology: Science fiction? Int. J. Surg. Pathol. 8, 261–263 (2000). PMID: 11494001
Hamilton, P.W., Wang, Y., McCullough, S.J.: Virtual microscopy and digital pathology in training and education. Apmis 120, 305–315 (2012)
Dandu, R.: Storage media for computers in radiology. Indian J. Radiol. Imag. 18, 287 (2008)
Reeder, M.M., Felson, B.: Gamuts in Radiology: Comprehensive Lists of Roentgen Differential Diagnosis. Pergamon Press (1977)
Goolsby, A.W., Olsen, L., McGinnis, M., Grossmann, C.: Clincial data as the basic staple of health learning - Creating and Protecting a Public Good. National Institute of Health (2010)
McDermott, J.E., Wang, J., Mitchell, H., Webb-Robertson, B.J., Hafen, R., Ramey, J., Rodland, K.D.: Challenges in biomarker discovery: combining expert insights with statistical analysis of complex omics data. Expert Opinion Med. Diagn. 7, 37–51 (2013)
Swan, A.L., Mobasheri, A., Allaway, D., Liddell, S., Bacardit, J.: Application of machine learning to proteomics data: Classification and biomarker identification in postgenomics biology. Omics-a J. Integr. Biol. 17, 595–610 (2013)
Jeanquartier, F., Jean-Quartier, C., Schreck, T., Cemernek, D., Holzinger, A.: Integrating open data on cancer in support to tumor growth analysis. In: Renda, M.E., Bursa, M., Holzinger, A., Khuri, S. (eds.) ITBAM 2016. LNCS, vol. 9832, pp. 49–66. Springer, Cham (2016). doi:10.1007/978-3-319-43949-5_4
Bleiholder, J., Naumann, F.: Data fusion. ACM Comput. Surv. (CSUR) 41, 1–41 (2008)
Lafon, S., Keller, Y., Coifman, R.R.: Data fusion and multicue data matching by diffusion maps. IEEE Trans. Pattern Anal. Mach. Intell. 28, 1784–1797 (2006)
Blanchet, L., Smolinska, A.: Data fusion in metabolomics and proteomics for biomarker discovery. In: Jung, K. (ed.) Statistical Analysis in Proteomics. MMB, vol. 1362, pp. 209–223. Springer, New York (2016). doi:10.1007/978-1-4939-3106-4_14
Hastie, T., Tibshirani, R., Friedman, J.: The Elements of Statistical Learning. Springer, New York (2009)
Bishop, C.M.: Pattern Recognition and Machine Learning (2006)
Burges, C.J.: A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Disc. 2, 121–167 (1998)
Kotropoulos, C., Pitas, I.: Segmentation of ultrasonic images using support vector machines. Pattern Recogn. Lett. 24, 715–727 (2003)
Ricci, E., Perfetti, R.: Retinal blood vessel segmentation using line operators and support vector classificatio. IEEE Trans. Med. Imaging 26, 1357–1365 (2007)
Orlando, J.I., Blaschko, M.: Learning fully-connected CRFs for blood vessel segmentation in retinal images. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014. LNCS, vol. 8673, pp. 634–641. Springer, Cham (2014). doi:10.1007/978-3-319-10404-1_79
Bauer, S., Nolte, L.-P., Reyes, M.: Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization. In: Fichtinger, G., Martel, A., Peters, T. (eds.) MICCAI 2011. LNCS, vol. 6893, pp. 354–361. Springer, Heidelberg (2011). doi:10.1007/978-3-642-23626-6_44
El-Naqa, I., Yang, Y., Wernick, M.N., Galatsanos, N.P., Nishikawa, R.M.: A support vector machine approach for detection of microcalcifications. IEEE Trans. Med. Imaging 21, 1552–1563 (2002)
Han, J.W., Breckon, T.P., Randell, D.A., Landini, G.: The application of support vector machine classification to detect cell nuclei for automated microscopy. Mach. Vis. Appl. 23, 15–24 (2012)
Breiman, L.: Random forests. Mach. Learn. 45, 4–32 (2001)
Criminisi, A., Jamie, S. (eds.): Decision Forests for Computer Vision and Medical Image Analysis. Springer, London (2013)
Zikic, D., Glocker, B., Konukoglu, E., Criminisi, A., Demiralp, C., Shotton, J., Thomas, O.M., Das, T., Jena, R., Price, S.J.: Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel MR. In: Ayache, N., Delingette, H., Golland, P., Mori, K. (eds.) MICCAI 2012. LNCS, vol. 7512, pp. 369–376. Springer, Heidelberg (2012). doi:10.1007/978-3-642-33454-2_46
Glocker, B., Pauly, O., Konukoglu, E., Criminisi, A.: Joint classification-regression forests for spatially structured multi-object segmentation. In: Fitzgibbon, A., Lazebnik, S., Perona, P., Sato, Y., Schmid, C. (eds.) ECCV 2012. LNCS, vol. 7575, pp. 870–881. Springer, Heidelberg (2012). doi:10.1007/978-3-642-33765-9_62
Richmond, D., Kainmueller, D., Glocker, B., Rother, C., Myers, G.: Uncertainty-driven forest predictors for vertebra localization and segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9349, pp. 653–660. Springer, Cham (2015). doi:10.1007/978-3-319-24553-9_80
Criminisi, A.: Anatomy detection and localization in 3D medical images. In: Criminisi, A., Shotton, J. (eds.) Decision Forests for Computer Vision and Medical Image Analysis. Advances in Computer Vision and Pattern Recognition. Springer, London (2013)
Štern, D., Ebner, T., Urschler, M.: From local to global random regression forests: exploring anatomical landmark localization. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 221–229. Springer, Cham (2016). doi:10.1007/978-3-319-46723-8_26
Štern, D., Ebner, T., Urschler, M.: Automatic localization of locally similar structures based on the scale-widening random regression forest. In: IEEE International Symposium on Biomedical Imaging (2017)
Hebb, D.: The Organization of Behavior. Wiley, New York (1949)
McCulloch, W.S., Pitts, W.: A logical calculus of the ideas immanent in nervous activity. Bull. Mathe. Biophys. 5, 115–133 (1943)
LeCun, Y., Bottou, L., Bengio, Y., Haffner, P.: Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2324 (1998)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet Classification with Deep Convolutional Neural Networks. In: Advances in Neural Information Processing Systems (2012)
Singh, D., Merdivan, E., Psychoula, I., Kropf, J., Hanke, S., Geist, M., Holzinger, A.: Human activity recognition using recurrent neural networks. In: Holzinger, A., Kieseberg, P., Tjoa, A.M., Weippl, E. (eds.) CD-MAKE 2017. LNCS, vol. 10410, pp. 267–274. Springer, Cham (2017). doi:10.1007/978-3-319-66808-6_18
Bengio, Y., Courville, A., Vincent, P.: Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 35, 1798–1828 (2013)
Liu, Y., Gadepalli, K., Norouzi, M., Dahl, G.E., Kohlberger, T., Boyko, A., Venugopalan, S., Timofeev, A., Nelson, P.Q., Corrado, G.S., Hipp, J.D., Peng, L., Stumpe, M.C.: Detecting cancer metastases on gigapixel pathology images. arXiv: 1703.02442 (2017)
Esteva, A., Kuprel, B., Novoa, R.A., Ko, J., Swetter, S.M., Blau, H.M., Thrun, S.: Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Proceedings Medical Image Computing and Computer-Assisted Intervention (2015)
Pan, S.J., Yang, Q.: A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2010)
Cai, J., Lu, L., Xie, Y., Xing, F., Yang, L.: Pancreas segmentation in MRI using graph-based decision fusion on convolutional neural networks. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 674–682. Springer, Cham (2017). doi:10.1007/978-3-319-66179-7_77
Payer, C., Štern, D., Bischof, H., Urschler, M.: Regressing heatmaps for multiple landmark localization using CNNs. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 230–238. Springer, Cham (2016). doi:10.1007/978-3-319-46723-8_27
Rozantsev, A., Lepetit, V., Fua, P.: On rendering synthetic images for training an object detector. Comput. Vis. Image Underst. 137, 24–37 (2015)
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., Courville, A., Bengio, Y.: Generative adversarial nets. In: Advances in Neural Information Processing Systems (2014)
Nie, D., Trullo, R., Petitjean, C., Ruan, S., Shen, D.: Medical image synthesis with context-aware generative adversarial networks. arXiv:1612.05362 (2016). Accepted MICCAI’17
Schmidhuber, J.: Deep learning in neural networks: An overview. Neural Netw. 61, 85–117 (2015)
Goodfellow, I., Bengio, Y., Courville, A.: Deep Learning. MIT Press (2016)
Malle, B., Kieseberg, P., Schrittwieser, S., Holzinger, A.: Privacy aware machine learning and the right to be forgotten. ERCIM News (Special Theme: Machine Learning) 107, 22–23 (2016)
Fosch Villaronga, E., Kieseberg, P., Li, T.: Humans forget, machines remember: Artificial intelligence and the right to be forgotten. Computer Security Law Review (2017)
Malle, B., Giuliani, N., Kieseberg, P., Holzinger, A.: The more the merrier - federated learning from local sphere recommendations. In: Holzinger, A., Kieseberg, P., Tjoa, A.M., Weippl, E. (eds.) CD-MAKE 2017. LNCS, vol. 10410, pp. 367–373. Springer, Cham (2017). doi:10.1007/978-3-319-66808-6_24
Dwork, C.: Differential privacy: a survey of results. In: Agrawal, M., Du, D., Duan, Z., Li, A. (eds.) TAMC 2008. LNCS, vol. 4978, pp. 1–19. Springer, Heidelberg (2008). doi:10.1007/978-3-540-79228-4_1
Kieseberg, P., Hobel, H., Schrittwieser, S., Weippl, E., Holzinger, A.: Protecting anonymity in data-driven biomedical science. In: Holzinger, A., Jurisica, I. (eds.) Interactive Knowledge Discovery and Data Mining in Biomedical Informatics. LNCS, vol. 8401, pp. 301–316. Springer, Heidelberg (2014). doi:10.1007/978-3-662-43968-5_17
Schrittwieser, S., Kieseberg, P., Echizen, I., Wohlgemuth, S., Sonehara, N., Weippl, E.: An algorithm for k-anonymity-based fingerprinting. In: Shi, Y.Q., Kim, H.-J., Perez-Gonzalez, F. (eds.) IWDW 2011. LNCS, vol. 7128, pp. 439–452. Springer, Heidelberg (2012). doi:10.1007/978-3-642-32205-1_35
Kieseberg, P., Schrittwieser, S., Mulazzani, M., Echizen, I., Weippl, E.: An algorithm for collusion-resistant anonymization and fingerprinting of sensitive microdata. Electron. Markets 24, 113–124 (2014)
Haerder, T., Reuter, A.: Principles of transaction-oriented database recovery. ACM Comput. Surv. (CSUR) 15, 287–317 (1983)
Bayer, R., McCreight, E.: Organization and maintenance of large ordered indexes. In: Broy, M., Denert, E. (eds.) Software Pioneers, pp. 245–262. Springer, Heidelberg (2002)
Fruhwirt, P., Kieseberg, P., Weippl, E.: Using internal MySQL/InnoDB B-tree index navigation for data hiding. In: Peterson, G., Shenoi, S. (eds.) DigitalForensics 2015. IAICT, vol. 462, pp. 179–194. Springer, Cham (2015). doi:10.1007/978-3-319-24123-4_11
Kieseberg, P., Schrittwieser, S., Mulazzani, M., Huber, M., Weippl, E.: Trees cannot lie: Using data structures for forensics purposes. In: Intelligence and Security Informatics Conference (EISIC), 2011 European, pp. 282–285. IEEE (2011)
Pantazos, K., Lauesen, S., Lippert, S.: De-identifying an EHR database-Anonymity, correctness and readability of the medical record. Stud. Health Technol. Inf. 169, 862–866 (2011)
Neamatullah, I., Douglass, M.M., Lehman, L.W.H., Reisner, A., Villarroel, M., Long, W.J., Szolovits, P., Moody, G.B., Mark, R.G., Clifford, G.D.: Automated de-identification of free-text medical records. BMC Med. Inform. Decis. Mak. 8, 32 (2008)
Al-hegami, A.S.: A biomedical named entity recognition using machine learning classifiers and rich feature set. Int. J. Comput. Sci. Netw. Secur. 17, 170–176 (2017)
Settles, B.: Biomedical named entity recognition using conditional random fields and rich feature sets. In: International Joint Workshop on Natural Language Processing in Biomedicine and its Applications, pp. 104–107 (2004)
Mavromatis, G.: Biomedical named entity recognition using neural networks 2015, 1–9 (2015)
Goldberg, Y., Levy, O.: word2vec explained: deriving Mikolov et al. negative-sampling word-embedding method. arXiv preprint arXiv:1402.3722 (2014)
Sweeney, L.: k-anonymity: A model for protecting privacy. Int. J. Uncertainty, Fuzziness and Knowl.-Based Syst. 10, 557–570 (2002)
Machanavajjhala, A., Gehrke, J., Kifer, D., Venkitasubramaniam, M.: l-diversity: Privacy beyond k-anonymity. In: Proceedings of the 22nd International Conference on Data Engineering, ICDE 2006, p. 24. IEEE (2006)
Li, N., Li, T., Venkatasubramanian, S.: t-closeness: Privacy beyond k-anonymity and l-diversity. In: IEEE 23rd International Conference on Data Engineering, ICDE 2007, pp. 106–115. IEEE (2007)
Nergiz, M.E., Atzori, M., Clifton, C.: Hiding the presence of individuals from shared databases. In: Proceedings of the 2007 ACM SIGMOD International Conference on Management of Data, pp. 665–676. ACM (2007)
Wong, R.C.W., Li, J., Fu, A.W.C., Wang, K.: (\(\alpha \), k)-anonymity: an enhanced k-anonymity model for privacy preserving data publishing. In: Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 754–759. ACM (2006)
Campan, A., Truta, T.M.: Data and structural k-Anonymity in social networks. In: Bonchi, F., Ferrari, E., Jiang, W., Malin, B. (eds.) PInKDD 2008. LNCS, vol. 5456, pp. 33–54. Springer, Heidelberg (2009). doi:10.1007/978-3-642-01718-6_4
Malle, B., Kieseberg, P., Weippl, E., Holzinger, A.: The right to be forgotten: towards machine learning on perturbed knowledge bases. In: Buccafurri, F., Holzinger, A., Kieseberg, P., Tjoa, A.M., Weippl, E. (eds.) CD-ARES 2016. LNCS, vol. 9817, pp. 251–266. Springer, Cham (2016). doi:10.1007/978-3-319-45507-5_17
Malle, B., Kieseberg, P., Holzinger, A.: DO NOT DISTURB? classifier behavior on perturbed datasets. In: Holzinger, A., Kieseberg, P., Tjoa, A.M., Weippl, E. (eds.) CD-MAKE 2017. LNCS, vol. 10410, pp. 155–173. Springer, Cham (2017). doi:10.1007/978-3-319-66808-6_11
Rafique, A., Azam, S., Jeon, M., Lee, S.: Face-deidentification in images using restricted boltzmann machines. In: ICITST, pp. 69–73 (2016)
Chi, H., Hu, Y.H.: Face de-identification using facial identity preserving features. In: 2015 IEEE Global Conference on Signal and Information Processing, GlobalSIP 2015, pp. 586–590 (2016)
Yu, F., Fienberg, S.E., Slavković, A.B., Uhler, C.: Scalable privacy-preserving data sharing methodology for genome-wide association studies. J. Biomed. Inform. 50, 133–141 (2014)
Simmons, S., Sahinalp, C., Berger, B.: Enabling privacy-preserving GWASs in heterogeneous human populations. Cell Syst. 3, 54–61 (2016)
Im, H.K., Gamazon, E.R., Nicolae, D.L., Cox, N.J.: On sharing quantitative trait GWAS results in an era of multiple-omics data and the limits of genomic privacy. Am. J. Hum. Genet. 90, 591–598 (2012)
Knoppers, B.M., Dove, E.S., Litton, J.E., Nietfeld, J.J.: Questioning the limits of genomic privacy. Am. J. Hum. Genet. 91, 577–578 (2012)
Aggarwal, C.C., Li, Y., Philip, S.Y.: On the hardness of graph anonymization. In: 2011 IEEE 11th International Conference on Data Mining (ICDM), pp. 1002–1007. IEEE (2011)
Baxter, J.: A model of inductive bias learning. J. Artif. Intell. Res. 12, 149–198 (2000)
Evgeniou, T., Pontil, M.: Regularized multi-task learning. In: Proceedings of the Tenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 109–117. ACM (2004)
Weinberger, K.Q., Saul, L.K.: Distance metric learning for large margin nearest neighbor classification. J. Mach. Learn. Res. 10, 207–244 (2009)
Parameswaran, S., Weinberger, K.Q.: Large margin multi-task metric learning. In: Lafferty, J., Williams, C., Shawe-Taylor, J., Zemel, R., Culotta, A. (eds.) Advances in Neural Information Processing Systems 23 (NIPS 2010), pp. 1867–1875 (2010)
McCloskey, M., Cohen, N.J.: Catastrophic interference in connectionist networks: The sequential learning problem. In: Bower, G.H. (ed.) The Psychology of Learning and Motivation, vol. 24, pp. 109–164. Academic Press, San Diego (1989)
French, R.M.: Catastrophic forgetting in connectionist networks. Trends Cogn. Sci. 3, 128–135 (1999)
Goodfellow, I.J., Mirza, M., Xiao, D., Courville, A., Bengio, Y.: An empirical investigation of catastrophic forgeting in gradient-based neural networks. arXiv:1312.6211v3 (2015)
Taylor, M.E., Stone, P.: Transfer learning for reinforcement learning domains: A survey. J. Mach. Learn. Res. 10, 1633–1685 (2009)
Sycara, K.P.: Multiagent systems. AI Mag. 19, 79 (1998)
Lynch, N.A.: Distributed Algorithms. Morgan Kaufmann, San Francisco (1996)
DeGroot, M.H.: Reaching a consensus. J. Am. Stat. Assoc. 69, 118–121 (1974)
Benediktsson, J.A., Swain, P.H.: Consensus theoretic classification methods. IEEE Trans. Syst. Man Cybern. 22, 688–704 (1992)
Weller, S.C., Mann, N.C.: Assessing rater performance without a gold standard using consensus theory. Med. Decis. Making 17, 71–79 (1997)
Olfati-Saber, R., Fax, J.A., Murray, R.M.: Consensus and cooperation in networked multi-agent systems. Proc. IEEE 95, 215–233 (2007)
Roche, B., Guegan, J.F., Bousquet, F.: Multi-agent systems in epidemiology: a first step for computational biology in the study of vector-borne disease transmission. BMC Bioinf. 9 (2008)
Kok, J.R., Vlassis, N.: Collaborative multiagent reinforcement learning by payoff propagation. J. Mach. Learn. Res. 7, 1789–1828 (2006)
Acknowledgments
We are grateful for valuable comments from the international reviewers.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Holzinger, A. et al. (2017). Machine Learning and Knowledge Extraction in Digital Pathology Needs an Integrative Approach. In: Holzinger, A., Goebel, R., Ferri, M., Palade, V. (eds) Towards Integrative Machine Learning and Knowledge Extraction. Lecture Notes in Computer Science(), vol 10344. Springer, Cham. https://doi.org/10.1007/978-3-319-69775-8_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-69775-8_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-69774-1
Online ISBN: 978-3-319-69775-8
eBook Packages: Computer ScienceComputer Science (R0)