Abstract
Accurate diagnosis of high-risk benign breast lesions is crucial in patient management since they are associated with an increased risk of invasive breast cancer development. Since it is not yet possible to identify the occult cancer patients without surgery, this limitation leads to retrospectively unnecessary surgeries. In this paper, we present a computational pathology pipeline for histological diagnosis of high-risk benign breast lesions from whole slide images (WSIs). Our pipeline includes WSI stain color normalization, ductal regions of interest (ROIs) segmentation, and cytological and architectural feature extraction to classify ductal ROIs into triaged high-risk benign lesions. We curated 93 WSIs of breast tissues containing high-risk benign lesions based on pathology reports and collected ground truth annotations from three different pathologists for the ductal ROIs segmented by our pipeline. Our method has comparable performance to a pool of expert pathologists.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Breast lesions
- Atypical ductal hyperplasia
- Computational pathology
- Pattern recognition
- Architectural pattern
- Classification
1 Introduction
Benign breast lesions are an important source of disagreement and uncertainty for pathologists when evaluating breast core biopsies as part of multidisciplinary breast cancer screening programs [6]. These benign lesions can be categorized into three groups: nonproliferative, proliferative without atypia, or atypical hyperplasia. Among these, atypical hyperplasias have a substantially elevated (approximately 4-fold) risk of breast cancer development [5]. Atypical hyperplasias, which include atypical ductal hyperplasia (ADH) and atypical lobular hyperplasia (ALH), are found in 12–17% of biopsies performed. More recently flat epithelial atypia (FEA), which is an alteration of the breast lobules, is defined as an additional type of atypical lesion with uncertain long term breast cancer risk [12]. Although this may change, FEA in a core biopsy is generally followed by excisional biopsy [2]. On the other hand, columnar cell change (CCC) is a relatively common, non-atypical proliferative lesion that is generally regarded as very low risk despite morphological similarity to FEA [11]. In this study, we focus on differentiating high-risk (ADH, FEA) vs. low-risk (CCC, normal duct) breast lesions.
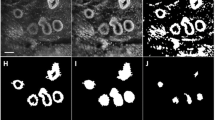
Sample ductal ROIs representing (A) atypical ductal hyperplasia (ADH), (B) flat epithelial atypia (FEA), (C) columnar cell change (CCC), and (D) normal duct. (E)–(H) Visualization of architectural patterns discovered in sample ROIs. Patterns are derived from a combination of cytological and architectural features and visualized by color coded objects (see x-axis of panel (I)). Note the overexpression of pattern #5 in ADH, #7 in FEA, and #15 in normal ducts (E)–(H). This observation is further supported by the histogram in panel (I), where we measure relative proportions of architectural patterns separately in each one of the categories: ADH, FEA, CCC, and normal.
Diagnostic criteria for high-risk benign lesions exist but rely on atypia, which is a subjective feature that may lack reproducibility, especially among non-subspecialist pathologists. Figure 1A–D show sample ROIs from breast lesions. ADH (Fig. 1A) is difficult because it can have overlapping features with later invasive breast cancer development. Widely accepted criteria for diagnosing ADH include: (1) atypical cell features, (2) architectural patterns, and (3) size or extent of the lesion. However, the first two criteria can be subjective or variable, making distinction from other cases problematic. FEA (Fig. 1B) generally refers to open (or rounded) ducts lined by disorganized arrays of atypical appearing cells including monomorphic appearance. They lack the orderly columnar arrangement seen in CCC (Fig. 1C), where ducts are also often open and rounded, but are lined by non-atypical cells that have a columnar arrangement. FEA and CCC can be difficult to distinguish as they have similar architecture (i.e. flat), and one must rely upon atypia.
To date, improved reproducibility and more consistent application of diagnostic criteria have been difficult to achieve for borderline cases such as ADH and FEA [6]. To begin quantifying diagnostic criteria, we have constructed a computational pathology pipeline for detecting high-risk benign breast lesions from whole slide images (WSIs). Although there are several studies on cancer detection in breast tissue images, to the best of our knowledge, our proposed pipeline is the first of its kind in detecting high-risk benign breast lesions. Previous studies [3, 4, 9, 15] used manually selected ROIs from WSIs to classify breast lesions. The approaches in [3, 4, 9] were based on cytological features, such as identifying and characterizing morphology and texture of nuclei. In [9], the authors combine both cytological and architectural features to demonstrate the importance of spatial statistics in separating cancer lesions from non-cancerous ones. Recently, an end-to-end system for detecting ductal carcinoma in situ (DCIS) was proposed by [1], in which ROIs from WSIs were delineated and classified into benign vs. DCIS. Their study explicitly excluded slides containing ADH due to high level of disagreement and the difficulty in collecting ground truth.
Our paper is the first attempt in building an end-to-end high-risk benign breast lesion detector for WSIs that includes WSI stain color normalization, ductal ROI segmentation, cytological and architectural feature extraction, and ductal ROI classification. A key contribution of this study is to encode morphometric properties of nuclear atypia (cytological) and combine them with the spatial distribution of the nuclei in relationship to stroma and lumen (architectural). Additionally, we collected high-risk benign lesion data and the ground truth annotations from three expert pathologists.
2 Methodology
2.1 Stain Color Normalization
Histopathology images can have a wide range of color appearances due to biological differences, slide preparation techniques, and imaging hardware. One way to reduce this variability is to preprocess the digital tissue images using color normalization methods. For the datasets that we collected for this project, we observed that previous methods [7, 18] either do not scale well to WSIs or generate artifacts such as blue backgrounds. To resolve these issues, the authors in [10] developed a scalable color normalization method based on opponent color spaces and a fast sampling-based strategy for parameter estimation. In particular, the color space is similar to HSV and is optimized for separating hematoxylin and eosin stains. Because this color space is angular, the stains are separated using a mixture of von Mises distributions. After separating the stains, the statistics of the source image is matched to a reference image up to the fourth order (Fig. 2A), a common method in texture synthesis. In addition, this method is scaled to work with large WSIs by an efficient sampling-based strategy for estimating von Mises parameters. This method has been evaluated by a comprehensive set of quantitative and qualitative performance measures and showed significant improvement over the state–of–the–art color normalization methods.
2.2 Ductal ROI Segmentation
To segment ductal regions of interest the authors in [10] observed that the spatial density of epithelial nuclei can be efficiently used to partition ducts from breast WSIs. In particular, the WSI is decomposed into superpixels [17] to approximately denote the nuclei, stroma, and lumen components of the tissue (Fig. 2B). Delaunay triangulation is performed on superpixel centers and a neighborhood graph is constructed for the entire WSI. The triangulation preserves physical distances and helps avoid the problem of connecting a fibroblast nucleus with an epithelial nucleus when they are separated by a large area of stroma. As a first approximation to the spatial density of the nuclei, neighboring superpixels are connected by an edge if their physical distance is under a threshold as given by the median distance between pairs of neighboring nuclei. A greedy connected component analysis is then run on this graph to cluster the superpixels into ROIs. Since the goal is to segment ductal ROIs, lumen superpixels are also clustered into ROIs and then merged with nuclei ROIs if they overlap.
2.3 Cytological Phenotyping
For the sake of phenotyping, we generate a more precise set of nuclei masks in each ductal ROI using Fiji [13]. We apply a simple threshold on hematoxylin color channel and obtain putative nuclei regions. We use watershed to separate touching and overlapping nuclei and used morphological operations to fill any remaining holes. Next, we eliminate small and large segmented objects and those near the image border. Finally, one round of erosion followed by dilation is performed to smoothen the nuclei shape.
To compute cytological phenotypes, we compute nuclear features as defined in [3]. There are 196 features including morphological features such as roundness, aspect ratio, bounding box dimensions; intensity features such as means, variance, skewness, and kurtosis in multiple color channels (RGB, HSV, La\(^*\)b\(^*\)); and texture features such as Haralick’s features and graph run length features for each nuclei. We observe three dominant phenotypes in this data, which we capture using Matlab’s \(k{-}\)means clustering algorithm, with \(k{-}\)means\(++\) as smart initialization and a warm start option. The three dominant phenotypes may be a consequence of normal, atypical, and pleomorphic nuclei in high-risk benign breast lesions (Fig. 2C). In addition, for the task of high-risk vs. low-risk classification, we construct a cytological feature (CF) vector for each ROI, which is a summary statistic (e.g., mean, median, std-dev, etc.) of the aforementioned 196 measures.
2.4 Architectural Phenotyping
For the sake of architectural phenotyping, we follow the idea presented in [16] to capture spatial properties of the tissue content. Mainly, the ROI is represented by 5 different objects: three cytologically phenotyped nuclei (\(\text {nuclei}_1\), \(\text {nuclei}_2\), \(\text {nuclei}_3\)) and two superpixel based components (stroma and lumen) as shown in Fig. 2C. To characterize the neighborhood around each object, a spatial network is constructed by breadth-first traversal from each object for a small number of depth levels (Fig. 2D). At each depth level we compute the probabilities of finding 15 different object connections (e.g., \(\text {nuclei}_1\)–\(\text {nuclei}_1\), \(\text {nuclei}_1\)–stroma, \(\text {nuclei}_1\)–lumen, etc.). As a result, for a maximum depth of 5, we generate a set of 75 probability values describing the neighborhood statistics for each object. The depth is set to a small number because the ductal ROIs are local and the breadth-first quickly covers its content.
To phenotype the spatial networks, we cluster the neighborhood statistics into q clusters by noting the principal subspace that captures \(95\%\) of the input variance. The architectural phenotypes are learned from applying k-NN algorithm. Each image is then represented by the relative proportion of q architectural patterns. We construct architectural feature vectors for three different scenarios based on (i) color based architectural features (AF–C) that use superpixel derived nuclei, stroma and lumen objects; (ii) cytologically phenotyped nuclei based architectural features (AF–N) that use nuclei phenotypes alone; and (iii) combined architectural features (AF–CN) that use nuclei phenotypes in combination with stromal and lumen superpixels.
3 Experiments and Results
3.1 Dataset
The pathological grading of cases was obtained from diagnostic pathology reports and was validated by our expert, who processed them a second time under light microscope. Whole slide images were then scanned using Aperio ScanScope XT at 0.5 microns per pixel resolution at \(20{\times }\) magnification. We collected a cohort of 46 ADH cases from a local hospital. These cases had a total of 269 WSIs, 93 of which were selected by the most experienced pathologist (P1) as containing at least one high-risk benign lesion.
From these 93 WSIs, 1759 ROIs were derived using the process in Sect. 2.2. Only 1009 ROIs are analyzed again by P1, forming the training set, and 750 ROIs analyzed by three expert pathologists (P1, P2, and P3), forming the test set. Each ROI could be classified as “ADH”, “columnar”, “flat epithelial”, “normal duct”, “don’t know”, and “other”. Any ROI classified as “don’t know” or “other” was discarded, leaving 839 ROIs in the training set. Any ROIs in the test set in which all three pathologists disagreed were discarded, leaving approximately 608 ROIs. From this assignment, “ADH” and “flat epithelial” are reclassified as “high-risk”, and “columnar” and “normal duct” are reclassified as “low-risk.” In total, we observe 251 “high-risk” and 588 “low-risk” in the training set and 71 “high-risk” and 537 “low-risk” in the test set. The dataset is highly imbalanced due to the low naturally rate of occurrence of ADH and FEA.
Within the test set, only 4% of the ROIs contain unanimous “high-risk” labels from the expert pathologists, 12% of the ROIs had at least two expert pathologists label it as “high-risk”, and 21% of the ROIs had at least one expert pathologist label it as “high-risk”. The overall Fleiss’ kappa score is calculated as .55, indicating a moderate agreement between the pathologists.
3.2 Results
Table 1 provides a summary of the results of recall over the high-risk ROIs and the weighted F-measure for both classes. We use these performance metrics because we are most interested in recognizing as many instances of high-risk ROIs while limiting false positives. Each of the three pathologists were asked to label each ROI, and their average performance informs the single expert pathologist baseline. All architectural feature sets (AF–C, AF–N, and AF–CN) were tested with Logistic Regression, Random Forest Walk, and SVM with SMOTE and cross-validation parameter scanning (which performed the best in all cases). From the original 196 cytological features, we generated 3530 summary statistics and performed feature selection, which provided 151 remaining features. This reduced set was tested with Naive Bayes, Decision Tree, SVM, and Logistic Regression (which performed the best). For completeness, we report results using Alexnet [8] and Overfeat [14]. ROI images were rescaled to \(512 \times 512\) and augmented the dataset using three rotations and two reflection. We loaded rebalanced batches and trained our nets for 2,000 epochs.
We find that using cytological features (CF) performs the best, but any architectural feature set performs similarly in both recall and F–measure. For both architectural and cytological features, we find that these feature sets outperform the majority classification and perform comparably to the average single expert pathologist classification with average computation time \(\tilde{2}6\) min on a single 2.4 GHz processor.
4 Conclusion and Discussions
Our goal in this paper was to build an end-to-end computational pipeline for histological diagnoses of high-risk vs. low-risk benign breast lesions. We used both cytological and architectural features in our method AF–CN. Figure 3 shows examples of AF–CN correctly and incorrectly classifying high-risk vs. low-risk ductal ROIs. We observe that the ROIs in these examples were correctly segmented by our pipeline. Figure 3A shows a typical example of ADH, where the roundness and monomorphism of nuclei are correctly captured by AF–CN (true positive). The high density of small ducts in Fig. 3B appears to be described by the architectural phenotyping of AF–CN, which correctly classified it as low-risk (true negative). However, the nuclei segmentation step falsely excludes overlapping nuclei, an indicator of low-risk lesions, which may factor into the incorrect high-risk classification of Fig. 3C (false positive). Finally, AF–CN misclassifies Fig. 3D as low-risk (false negative) possibly due to the AF–CN’s insufficient characterization of the shape properties of lumen regions. In our study, we observe that our combinations of cytological and architectural features did not provide the expected improvement over using only cytological features. The examples in Fig. 3 suggest that we can improve the performance by extracting additional architectural phenotypes. Our results highlight the challenge of diagnosing atypical breast lesions. Effective computational pathology pipelines rely on ground truth information, but this was surprisingly elusive in our study. It is very likely that a combined approach of more specimens (i.e. more ADH or FEA examples), larger numbers of pathologists, and perhaps consensus decisions would improve the reliability of ground truth. Regardless, our approach enables us to begin understanding what the exact “atypical” features are; this may permit future pipelines to better determine truly high-risk lesions and may also permit retraining of pathologists to understand what these features might be.
References
Bejnordi, B., et al.: Automated detection of DCIS in whole-slide H&E stained breast histopathology images. IEEE-TMI 35(9), 2141–2150 (2016)
Calhoun, B., et al.: Management of flat epithelial atypia on breast core biopsy may be individualized based on correlation with imaging studies. Mod. Pathol. 28(5), 670–676 (2015)
Dong, F., et al.: Computational pathology to discriminate benign from malignant intraductal proliferations of the breast. PLoS One 9(12), e114885 (2014)
Dundar, M., et al.: Computerized classification of intraductal breast lesions using histopathological images. IEEE-TBE 58(7), 1977–1984 (2011)
Dupont, W., Page, D.: Risk factors for breast cancer in women with proliferative breast disease. N. Engl. J. Med. 312(3), 146–151 (1985)
Elmore, J., et al.: Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA 313(11), 1122–1132 (2015)
Khan, A., et al.: A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE-TBE 61(6), 1729–1738 (2014)
Krizhevsky, et al.: Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1097–1105 (2012)
Nguyen, L., et al.: Architectural patterns for differential diagnosis of proliferative breast lesions from histopathological images. In: IEEE-ISBI (2017)
Nguyen, L., et al.: Spatial statistics for segmenting histological structures in H&E stained tissue images. IEEE-TMI PP(99), 1 (2017)
Pinder, S., Reis-Filho, J.: Non-operative breast pathology: columnar cell lesions. J. Clin. Pathol. 60(12), 1307–1312 (2007)
Said, S., et al.: Flat epithelial atypia and risk of breast cancer: a mayo cohort study. Cancer 121(10), 1548–1555 (2015)
Schindelin, J., et al.: Fiji: an open-source platform for biological-image analysis. Nat. Methods 9(7), 676–682 (2012)
Sermanet, P., et al.: Overfeat: Integrated recognition, localization and detection using convolutional networks. arXiv:1312.6229 (2013)
Srinivas, U., et al.: SHIRC: a simultaneous sparsity model for histopathological image representation and classification. In: IEEE-ISBI, pp. 1118–1121 (2013)
Tosun, A., Gunduz-Demir, C.: Graph run-length matrices for histopathological image segmentation. IEEE-TMI 30(3), 721–732 (2011)
Tosun, A., et al.: Object-oriented texture analysis for the unsupervised segmentation of biopsy images for cancer detection. Pattern Recogn. 42(6), 1104–1112 (2009)
Vahadane, A., et al.: Structure-preserving color normalization and sparse stain separation for histological images. IEEE-TMI 35(8), 1962–1971 (2016)
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Tosun, A.B. et al. (2017). Histological Detection of High-Risk Benign Breast Lesions from Whole Slide Images. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer-Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10434. Springer, Cham. https://doi.org/10.1007/978-3-319-66185-8_17
Download citation
DOI: https://doi.org/10.1007/978-3-319-66185-8_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66184-1
Online ISBN: 978-3-319-66185-8
eBook Packages: Computer ScienceComputer Science (R0)