Abstract
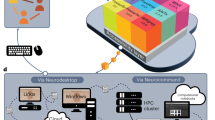
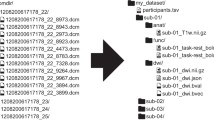
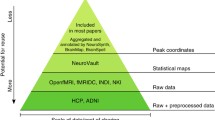
In the last decade, a large amount of neuroimaging datasets became publicly available on different archives, so there is an increasing need to manage heterogeneous data, aggregate and process them by means of large-scale computational resources. ReCaS datacenter offers the most important features to manage big datasets, process them, store results in efficient manner and make all the pipeline steps available for reproducible data analysis. Here, we present a scientific computing environment in ReCaS datacenter to deal with common problems of large-scale neuroimaging processing. We show the general architecture of the datacenter and the main steps to perform multidimensional neuroimaging processing.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Amoroso, N., et al.: Medical physics applications in Bari ReCaS farm. In: High Performance Scientific Computing Using Distributed Infrastructures: Results and Scientific Applications Derived from the Italian PON ReCaS Project, pp. 271–278. World Scientific (2017)
Behrens, T.E., Berg, H.J., Jbabdi, S., Rushworth, M.F., Woolrich, M.W.: Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? Neuroimage 34(1), 144–155 (2007)
Cox, R.W.: AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29(3), 162–173 (1996)
Craddock, C., et al.: Towards automated analysis of connectomes: the configurable pipeline for the analysis of connectomes (C-PAC). Front Neuroinform. 42 (2013)
Di Martino, A., et al.: The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry 19(6), 659 (2014)
Dinov, I., et al.: Efficient, distributed and interactive neuroimaging data analysis using the LONI pipeline. Front. Neuroinform. 3, 22 (2009)
Dinov, I.D., et al.: High-throughput neuroimaging-genetics computational infrastructure. Front. Neuroinform. 8, 41 (2014)
Fischl, B.: Freesurfer. Neuroimage 62(2), 774–781 (2012)
Gorgolewski, K.J., et al.: The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 160044 (2016)
Jack Jr., C.R., et al.: The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging: Off. J. Int. Soc. Magn. Reson. Med. 27(4), 685–691 (2008)
Jenkinson, M., Beckmann, C.F., Behrens, T.E., Woolrich, M.W., Smith, S.M.: FSL. Neuroimage 62(2), 782–790 (2012)
Lella, E., et al.: Communicability characterization of structural DWI subcortical networks in Alzheimer’s disease. Entropy 21(5), 475 (2019)
Lella, E., et al.: Communicability disruption in Alzheimer’s diseaseconnectivity networks. J. Complex Netw. 7(1), 83–100 (2018)
Lombardi, A., et al.: Modelling cognitive loads in schizophrenia by means of new functional dynamic indexes. NeuroImage 195, 150–164 (2019)
Maglietta, R., et al.: Automated hippocampal segmentation in 3D MRI using random undersampling with boosting algorithm. Pattern Anal. Appl. 19(2), 579–591 (2016)
Penny, W.D., Friston, K.J., Ashburner, J.T., Kiebel, S.J., Nichols, T.E.: Statistical Parametric Mapping: The Analysis of Functional Brain Images. Elsevier, Amsterdam (2011)
Poldrack, R.A., Gorgolewski, K.J.: Making big data open: data sharing in neuroimaging. Nat. Neurosci. 17(11), 1510 (2014)
Tournier, J.D., Calamante, F., Connelly, A.: MRtrix: diffusion tractography in crossing fiber regions. Int. J. Imaging Syst. Technol. 22(1), 53–66 (2012)
Van Horn, J.D., Toga, A.W.: Human neuroimaging as a “big data” science. Brain Imaging Behav. 8(2), 323–331 (2014)
Zuo, X.N., et al.: An open science resource for establishing reliability and reproducibility in functional connectomics. Sci. Data 1, 140049 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Lombardi, A. et al. (2019). Multidimensional Neuroimaging Processing in ReCaS Datacenter. In: Montella, R., Ciaramella, A., Fortino, G., Guerrieri, A., Liotta, A. (eds) Internet and Distributed Computing Systems . IDCS 2019. Lecture Notes in Computer Science(), vol 11874. Springer, Cham. https://doi.org/10.1007/978-3-030-34914-1_44
Download citation
DOI: https://doi.org/10.1007/978-3-030-34914-1_44
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-34913-4
Online ISBN: 978-3-030-34914-1
eBook Packages: Computer ScienceComputer Science (R0)