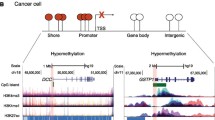
Abstract
DNA methylation is an epigenetic modification consisting in the addition of a methyl group to the position 5 of a cytosine in a CpG context. In normal mammalian cells, while CpG islands, mostly concentrated at promoter regions, are protected from DNA methylation, intergenic and repetitive regions are normally hypermethylated. In cancer cells, a massive change in the global methylation pattern occurs. Intergenic and repetitive regions of the genome become hypomethylated leading to the reactivation of transposable elements and genomic instability. In contrast, a focal hypermethylation of CpG islands at promoter regions occurs and it is normally associated to gene expression downregulation. Thus, aberrant DNA methylation is one of the most striking features of cancer cells and several studies have demonstrated that cancer-specific methylation patterns exist. For this reason, DNA methylation represents an extremely useful biomarker for several applications, including cancer risk definition, prediction of clinical outcomes, treatment response and cancer relapse. Finally, the association between DNA methylation and gene expression, although notoriously recognized, is not yet fully known, and the study of DNA methylation alterations in cancer and their consequences can help elucidate the mechanisms underlying this relationship.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Akiyama Y, Watkins N, Suzuki H et al (2003) GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol 23:8429–8439
Antonelli M, Fadda A, Loi E et al (2018) Integrated DNA methylation analysis identifies topographical and tumoral biomarkers in pilocytic astrocytomas. Oncotarget 9:13807–13821
Barault L, Amatu A, Siravegna G et al (2018) Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut 67:1995–2005
Baylin SB (2005) DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2:S4–S11
Baylin SB, Ohm JE (2006) Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6:107–116
Berdasco M, Esteller M (2010) Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell 19:698–711
Bindra RS, Glazer PM (2007) Co-repression of mismatch repair gene expression by hypoxia in cancer cells: role of the Myc/Max network. Cancer Lett 252:93–103
Bindra RS, Gibson SL, Meng A et al (2005) Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res 65:11597–11604
Church TR, Wandell M, Lofton-Day C et al (2014) Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 63:317–325
Eden A, Gaudet F, Waghmare A et al (2003) Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300:455–455
Fadda A, Gentilini D, Moi L et al (2018) Colorectal cancer early methylation alterations affect the crosstalk between cell and surrounding environment, tracing a biomarker signature specific for this tumor. Int J Cancer 143:907–920
Feigin ME (2013) Harnessing the genome for characterization of G-protein coupled receptors in cancer pathogenesis. FEBS J 280:4729–4738
Gal-Yam EN, Egger G, Iniguez L et al (2008) Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A 105:12979–12984
Greger V, Passarge E, Höpping W et al (1989) Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet 83:155–158
Hinoue T, Weisenberger DJ, Lange CPE et al (2012) Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 22:271–282
Howard G, Eiges R, Gaudet F et al (2008) Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 27:404–408
Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3:415–428
Jones PA, Baylin SB (2007) The epigenomics of cancer. Cell 128:683–692
Keshet I, Schlesinger Y, Farkash S et al (2006) Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet 38:149–153
Kinzler KW, Vogelstein B (1996) Lessons from hereditary colorectal cancer. Cell 87:159–170
Klett H, Balavarca Y, Toth R et al (2018) Robust prediction of gene regulation in colorectal cancer tissues from DNA methylation profiles. Epigenetics 13:386–397
Krishnamachary B, Zagzag D, Nagasawa H et al (2006) Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor–null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res 66:2725–2731
Lam K, Pan K, Linnekamp JF et al (2016) DNA methylation based biomarkers in colorectal cancer: a systematic review. Biochim Biophys Acta Rev Cancer 1866:106–120
Lee SH, Kim J, Kim W-H et al (2009) Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells. Oncogene 28:184–194
Levanon D, Bernstein Y, Negreanu V et al (2011) Absence of Runx3 expression in normal gastrointestinal epithelium calls into question its tumour suppressor function. EMBO Mol Med 3:593–604
Long C, Yin B, Lu Q et al (2007) Promoter hypermethylation of the RUNX3 gene in esophageal squamous cell carcinoma. Cancer Investig 25:685–690
Moarii M, Boeva V, Vert J-P et al (2015) Changes in correlation between promoter methylation and gene expression in cancer. BMC Genomics 16:873
Ohm JE, McGarvey KM, Yu X et al (2007) A stem cell–like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39:237–242
Oyer JA, Chu A, Brar S et al (2009) Aberrant epigenetic silencing is triggered by a transient reduction in gene expression. PLoS One 4:e4832
Phipps AI, Limburg PJ, Baron JA et al (2015) Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 148:77–87.e2
Rauch T, Li H, Wu X et al (2006) MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res 66:7939–7947
Rauch T, Wang Z, Zhang X et al (2007) Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A 104:5527–5532
Rodriguez J, Frigola J, Vendrell E et al (2006) Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res 66:8462–9468
Schlesinger Y, Straussman R, Keshet I et al (2007) Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39:232–236
Sharma S, Kelly TK, Jones PA (2010) Epigenetics in cancer. Carcinogenesis 31:27–36
Sproul D, Nestor C, Culley J et al (2011) Transcriptionally repressed genes become aberrantly methylated and distinguish tumors of different lineages in breast cancer. Proc Natl Acad Sci U S A 108:4364–4369
Sproul D, Kitchen RR, Nestor CE et al (2012) Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome Biol 13:R84
Timp W, Feinberg AP (2013) Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer 13:497–510
Viré E, Brenner C, Deplus R et al (2006) The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439:871–874
Widschwendter M, Fiegl H, Egle D et al (2007) Epigenetic stem cell signature in cancer. Nat Genet 39:157–158
Wong Doo N, Makalic E, Joo JE et al (2016) Global measures of peripheral blood-derived DNA methylation as a risk factor in the development of mature B-cell neoplasms. Epigenomics 8:55–66
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Loi, E., Zavattari, P. (2019). CpG Islands Methylation Alterations in Cancer: Functionally Intriguing Security Locks, Useful Early Tumor Biomarkers. In: Jurga, S., Barciszewski, J. (eds) The DNA, RNA, and Histone Methylomes. RNA Technologies. Springer, Cham. https://doi.org/10.1007/978-3-030-14792-1_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-14792-1_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-14791-4
Online ISBN: 978-3-030-14792-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)