Abstract
Research Data Management (RDM) receives more and more attention as a core component of scientific work. This importance equally stems from the scientific work with ever-increasing amounts of data, the value of this data for subsequent use, and the formal requirements of funding agencies. While these requirements are widely accepted among the research communities in general, the individual acceptance depends on many factors. In particular, we found that the ratio between the benefits achieved by RDM and the burdens imposed is not equal among the different roles that participate in the scientific process. In consequence, we analyse how we can optimize this ratio by different factors. Despite these different factors, common to all solutions is the demand for accessible and persistent storage that suits the particular needs imposed by RDM. At the Universities of Aachen, Bochum, Dortmund, Duisburg-Essen, and Cologne, we started a joint project to build up a distributed storage infrastructure dedicated to the needs of RDM and to address some of the acceptance factors.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Research data (RD) is the outcome as well as the foundation of scientific work. Researchers need an environment that enables them to work efficiently and securely with their research data (cf. [KE13, EU10]. National and international research funding institutions, such as the European Union program Horizon 2020, the German Research Foundation, the HRK, the German university rectors’ conference [Ho14, Ho15] and the Federal Ministry of Education and Research as well as various publishers (e.g. NATUREFootnote 1), are increasingly requiring scientists and scholars to plan and execute good data management practices. An obligation to archive produced data already exists by carrying out “good scientific practice” [RW11], some even ask for the publication of primary data, e.g. the Open Data Pilot of the EUFootnote 2.
To fulfill these growing requirements and thus make research data accessible and usable for subsequent research projects has become an inevitable objective for all scientific institutions. Thus, structures and processes have to be established to relieve the scientists from these tasks and let them focus their primary work. For this reason, there are many (inter-)nationally coordinated activities as well as activities at German federal state and local levels to tackle these questions. The coordinating activity in the state of Northrhine-Westphalia takes place in the context of “DH-NRW” (“Digitale Hochschule NRW” \( \Leftrightarrow \) “Digital University – Northrhine-Westphalia”). Here, activities at various levels, from communication and awareness-building activities, the compilation of a central, structured information repository about the requirements as well as of possible methods and tools scientists can use, up to detailed process charts to help scientists as well as central units within the universities to understand their mutual demands.
This contribution presents the approach of our universities to build a local infrastructure that integrates with these activities.
2 Requirements by Users
In many surveys and interviews [ES16], we tried to learn the users’ needs and requirements. All responses summed up, users primarily like to keep their current workflow (which is “best” supported by their local solution), and so any RDM infrastructure is expected to fit into whatever.
In essence, that means a user interface individually tailored at least to the requirements of a scientific community, ideally to a department’s habits. Such a level of customization currently does not seem a realistic requirement, so we need to establish ways to support the researchers by more standardized solutions.
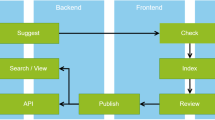
In order to draft such a solution – or a set of solutions – we first tried to translate the scientists’ requirements into processes and activities. At this level, RDM software should be easy to use and capable of supporting the scientists in managing their data. For the process level, we tried to model the various roles that participate within the scientific process (s. Fig. 1) in order to better structure and understand the scientists’ needs.
The most important roles are the Scientist and the Science-/project manager. Further roles are the society, i.e. anyone interested in results and data, and the external scientist.
The scientist generates data, makes annotations (i.e., generates metadata) and, at a later stage in a project, evaluates the data, tries models and parameters and so on. The science manager is involved in that evaluation process, too. Furthermore, he might manage several projects in parallel or in sequence, so he needs to be able to find and analyze data across several projects, a task often hindered by simple directory structures with implicit knowledge (sometimes buried in paper notebooks).
These various activities are not equally distributed over the lifecycle of research data.
The well-known domain model [KE13] visualized in Fig. 2 has proven to be a valuable means to a common understanding and basic structuring of research data management and the lifecycle of research data. In our context, it helps to structure demands and solutions according to their occurrence during the information life cycle. It distinguishes four domains where researchers act: the private domain, the group domain, the persistent domain, and the public domain enabling access and reuse. In our understanding, this domain model tries to capture the phases data and metadata undergo. The research process itself has much more loops and iterations like getting a set of data, evaluating, getting more data, perhaps with different experimental settings, and so on.
Domain model [KE13, ES16]
Research typically starts in the private domain where a researcher mainly works alone or in very small teams on his data. In most cases, the data is then with more collaborators, most often of other institutions or organizations. This phase is typically residing within the group domain. This wider sharing usually causes to add metadata well known to the individual researcher but essential to get an understanding by collaborators and thus has to be available with the shared data. Due to the duality of external collaborators being colleagues as well as competitors, identity based authentication and role based access control is important to participate in identity lifecycle management, role management etc. in order to have traceable access to data.
Once publications have been written or, at least, at the end of a project, good scientific practice and formal requirements asks for long-term storage of primary research data. At least the data that has led to the results of the project or the publication must be archived. Again, the need to capture further descriptive information rises since it cannot be ensured that people that have produced data will still be available once the data needs to be accessed. This is obvious for the public domain. Using the data by other researchers requires a thorough understanding based on an encompassing documentation with comprehensive and as far as possible standardized metadata.
The previous analyses lead to the impression that the value and benefit of RDM for the organization, for society, and for the project manager are very clear while the individual benefit for a scientist is not as obvious. We further followed this idea and tried to name the benefits and burdens, sorted by the domains of the domain model and by role (s. Table 1). Here, we split up the Project Manager role – often being a university professor – into the role of the principal investigator within a scientific context, the role of the head/manager of his department, and, eventually, the role of the organization’s head. With this structure, Table 1 shows a sketch of benefits (denoted by “+”) and burdens (“−”).
Despite the uncertainty of entries in this table, we see the workload to feed the RDM process lying mainly on the scientist where the individual benefit, in particular for scientists that leave the university after their graduation, appears to be marginal. On the other hand, a broad acceptance by the scientific community at our universities seems to be crucial for a successful implementation of RDM. Archives with just the mandatory fields filled to comply with the regulations appears to be too little to be useful.
With this background, we started our project with the aim to increase the benefit and to lower the burden for scientists.
3 Joint Project
When we look at the domain model (Fig. 2), an implicit assumption is that the “research” – i.e., the project – has already started somehow and that the infrastructure is aware of this project as well as of all other projects (Fig. 3).
However, besides the eventual project proposal and approval process, the incarnation of a project in an infrastructure at least has the potential of being a quite tedious task. After all interviews with researchers that is a strong reason why projects are not reflected in the technical and process infrastructure. On the other hand, as an institution we are highly to be able to differentiate each project since this gives a chance for any further treatment in a structured manner.
We therefore suggest to introduce a sort of a “0th domain” like depicted in Fig. 4. In particular, this domain should cover the technical instantiation of a project. Technically, we are going to build a portal that collects a project description, a timeframe and an estimation of the data to be expected.
We are going to ask for project proposals where the scientists or project managers can apply for storage on their own estimates. For the RD process, this yields the first metadata for the research data to come, namely the project identifier. Coherent with our aim to make things as easy as possible for our scientists we are planning a self-service portal that uses – at least up to a certain amount of data volume – AI to approve the project proposal. This approach promises to collect reasonable proposals and simultaneously an instant provisioning of storage resources, thereby lowering the burden to get storage quota approval.
With these (meta) data, we already know a lot about a project from an infrastructural perspective. Even more, the system underlying that portal implicitly can now form a “project lifecycle management”, delivers reports and so on.
As the next step, the researcher can form his or her project team by creating a group in the directory service and invited colleagues to this group as described in [EB13].
By integrating with other project related services, we can offer here stuff like a project web site, a groupware group for mails and appointments, a git project and so on.
Figure 4 also depicts the different infrastructure levels that support the data handling in the domains. The bar that runs along all the domains is labelled “Meta data”: Starting from the information collected during the project initialization in the “0th domain” we have to keep as much meta data as possible, either for the researcher himself, in order to support the evaluation of his experimental data, for team members or successors to make a use of the data and, when eventually published, to find and understand the data. The storage level in comparison in more segmented. Here we plan an online storage for the active project phase. After completion of a project or a thesis, the relevant data is put into an archive system. Currently, this is a dedicated tape based system, but the most important aspect here is that here long-term lifecycle management is handled like “keep at least for 10 years”.
Six Universities of the German state of North Rhine -Westphalia, Aachen (RWTH + Uni of applied Science), Bochum, Dortmund, Duisburg-Essen, and Cologne embarked to build modules and in particular the one named “online storage” as an infrastructure for RD-storage and RDM.
The RD-storage will be a 2-tier solution with an access layer as tier 1 and a persistency layer as tier 2. In that model, tier 1will offer access by NAS protocols and web/cloud protocols. In addition, community specific tools and portals are located in this tier to offer means to handle data and metadata and thus enable aspects of RDM. Common to all functions on tier 1 is a single authentication and authorization service that provides identities and identity based role information.
Tier 2 will be realized by a distributed object storage system. Due to the amount of “hot” research data at out universities, the total amount of storage will be in the 10 PB range. This tier addresses the above-mentioned “capacity, persistency, and security” aspect.
At each of our universities, we will realize this tiered configuration. The main advantages of the joint project is the co-operation at gaining expertise in running such a complex system in a way that it fits to the specific demand of RDM as well as in creating new solutions. Furthermore, we will use cross-site replication for protection of selected data.
As said, most important for our project is the goal to make RDM an integral part of every scientist’s workflow [RW11, RW16]. To reach this, an inevitable requirement to convince scientists is an availability very near 100%. This also requires an adequate availability of technical experts, which is much easier to accomplish by a larger – virtual – team.
A common question for all teams running storage infrastructure is how to manage the amount of data. There are techniques like billing for used space, setting quotas (by which rules ever) etc. Since we see the depicted RD storage infrastructure different from general-purpose storage but as a scientific resource, the “0th domain” will implement a process similar to the one which is established in the high performance computing regime since long and which matches perfectly with current funding regulations.
As said above, a project will start in the “0th domain”. Therefore, we expect to have a project id before data arrived. A side effect of this is that we can implement only project based storage quota and thereby avoid the difficulties that arise when mixing user quota with group (i.e., group) quota.
While technical parameters are necessary to be met, the scientists also ask for processes that support them doing science. Here, specific tools for every special field come in where the specialties how research data is handled, annotated, and used in this field are supported in a way that its use generates a value for the scientist. Due to the multitude of these tools, we are working closely together with researchers from different communities to implement a sufficient set of these.
The blocks named “RD user interface” will be systems that connect the scientists’ with their data beyond file access. We are planning a range from quite customizable web tools that allow managing meta data and connect it to data. We also see the opportunity for server based data analysis tools, either as PaaS or as SaaS. In any case, by being tied to one authentication service, all access methods on this layer will enforce consistent access rules.
To benefit from this, the logical interfaces must be the same across all sites, so besides technical co-operation we are also in the process of implementing matched change processes.
Common to all domains are the aspects of user-orientation or community orientation. The first means handling as well as the functional perspective, and integration into existing personal workflows. The second means the ability to integrate in the workflows that are established within a scientific community. This ability appears to be much more important than an institutional view.
This said, a strong focus of our RDM activities is about workflows, processes, metadata, and user interfaces.
However, every data has to be stored in a persistent and convenient manner and in a way, which fits the specifics of RDM. In particular, we see here the necessity to offer a broad range of protocols to access the storage by either legacy as well as modern, REST aware software. In our opinion, this is a prerequisite to offer services to the multitude of community specific software layers.
On the other hand, key aspects of RDM are the knowledge about the persons involved in the scientific workflow, and the ability to create cross references between stored data and metadata stored anywhere.
The cross references have to be inert against the specific location of the data in the storage hierarchy. This is achieved by storing the data in object storage and linking persistent, resolvable identifiers to the object id.
The knowledge about the persons start with the identity management systems established at our universities, relying on well-established methods of integrating another organization’s members in an identity management system [EB13]. However, since scientific collaboration depend on cross-university access, we have set up a concept for mapping the partner university’s users to each other’s user directory. In a next step, we are evaluating whether we can better support the scientific collaboration to have these resources linked to common scientific identity providers like Orchid or ResearcherID. This could pave the way to link directly a scientist’s results to his publication list and thus contributing to the individual benefit.
In general, we discuss data and their descriptions and annotations, i.e. their metadata. In this context, “data” is often understood as “original” data as obtained from experiments or simulations. However, a lot of research is made by using existing data, e.g. by applying a new analysis, a new way of combining information and so on. Regardless of the fact that this type of scientific work generates new data and metadata and results, these results rely on the previous generation of that data. Even with proper citation, it’s not trivial to keep track of such a dependency, in particular in cases when there are multiple layers of data dependency. Here, the blockchain technology could be a method to create a link chain for data citations, so that derived data can be tracked down to the original raw data even across multiple “hops”. Even more, starting from any point in that chain it allows finding dependent data. Due to the blockchain concept, such a link chain would even be institution agnostic.
As said, scientists and project managers have to rule with the regulations on RDM imposed by funding agencies etc. Here we see a way to support and thus lower the burden by taking (partial) responsibility in such a way that using the centrally supplied tools and infrastructure for research data will help the scientists to do things in a compliant way. Of course, that means an appropriate “Service level agreement” that goes significantly beyond the traditional “uptime and backup” promise.
4 Conclusion
In many cases, the “RDM process” is entered when the “real science” is done, namely when a project or a partial project has been completed and is due for archival and, eventually, for publication. In the common domain model, it is domains 3 and 4.
In our project, we are trying to “push” the entry point into RDM closer to the scientists’ workplace. From an institutional perspective, this improves the chance to get more data with better annotations as early as possible and thus allows protecting that data from being lost. From the individual scientists’ perspective, this approach has the downside that the daily work must be adopted to such a framework. However, is offers the great chance that the scientists have an individual benefit from using RDM. These benefits might stem from data analysis tools that use the aggregated metadata for better analyzing the data, from the safekeeping of data by the storage infrastructure, and by helping the scientist adhering to compliance rules.
Our next steps in that project are to implement the storage components and to combine software and APIs to use our growing RDM infrastructure as seamless as possible, where this will be a long-running project where the tool part will grow and develop incrementally as more and more scientists adopt to the RD process and articulate specific demands.
References
European Union: Riding the Wave. How Europe can gain from the rising tide of scientific data (2010). http://ec.europa.eu/information_society/newsroom/cf/document.cfm?action=display&doc_id=707. Last Access 11 Jan 2016
Hochschulrektorenkonferenz: Management of research data – a key strategic challenge for university management (2014). http://www.hrk.de/positionen/gesamtliste-beschluesse/position/convention/management-von-forschungsdaten-eine-zentrale-strategische-herausforderung-fuer-hochschulleitungen/. Last Access 12 Jan 2016
Hochschulrektorenkonferenz: How university management can guide the development of research data management. Orientation paths, options for action and scenarios (2015). http://www.hrk.de/positionen/gesamtliste-beschluesse/position/convention/wiehochschulleitungen-die-entwicklung-des-forschungsdatenmanagements-steuern-koennen-orientierung/. Last Access 12 Jan 2016
Eifert, T., Muckel, S., Schmitz, D.: Introducing Research Data Management as a Service Suite at RWTH Aachen University. GI Lecture Notes in Informatics – Proceedings, vol. P-257, pp. 55–64, ISSN (Print) 1617-5468, ISBN (Print) 978-3-88579-651-0
RWTH Aachen University: Grundsätze zur Sicherung guter wissenschaftlicher Praxis der Rheinisch-Westfälischen Technischen Hochschule Aachen. Amtliche Bekanntmachung vom, 11 January 2011. http://www.rwth-aachen.de/global/show_document.asp?id=aaaaaaaaaaaoyxb. Last Access 12 Jan 2016
RWTH Aachen University: Leitlinien zum Forschungsdatenmanagement für die RWTH Aachen. Rektoratsbeschluss vom, 08 March 2016. http://www.rwth-aachen.de/global/show_document.asp?id=aaaaaaaaaaqwpfe&download=1. Last Access 10 March 2016
Eifert, T., Bunsen, G.: Grundlagen und Entwicklung von Identity Management an der RWTH Aachen. PIK - Praxis der Informationsverarbeitung und Kommunikation. Band 36, Heft 2, Seiten 109–116 (2013). doi:10.1515/pik-2012-0053
Klar, J., Enke, H.: Rahmenbedingungen einer disziplinübergreifenden Forschungsdateninfrastruktur. Report Organisation und Struktur (2013). doi:10.2312/RADIESCHEN_005. Last Access 12 Jan 2016
Acknowledgements
A joint project group of our universities has carried out the depicted work. Our thanks go to J. Kather, B. Magrean, M. Politze, R. Reinecke, and M.S. Müller for the many fruitful discussions.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Eifert, T., Schilling, U., Bauer, HJ., Krämer, F., Lopez, A. (2017). Infrastructure for Research Data Management as a Cross-University Project. In: Yamamoto, S. (eds) Human Interface and the Management of Information: Supporting Learning, Decision-Making and Collaboration. HIMI 2017. Lecture Notes in Computer Science(), vol 10274. Springer, Cham. https://doi.org/10.1007/978-3-319-58524-6_39
Download citation
DOI: https://doi.org/10.1007/978-3-319-58524-6_39
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-58523-9
Online ISBN: 978-3-319-58524-6
eBook Packages: Computer ScienceComputer Science (R0)