Abstract
This study examined postmenstrual age (PMA) estimation (in weeks) from brain diffusion MRI of very preterm born infants (born <31weeks gestational age), with an objective to investigate how differences in estimated brain age and PMA were associated with the risk of Cerebral Palsy disorders (CP). Infants were scanned up to 2 times, between 29 and 46 weeks (w) PMA. We applied a deep learning 2D convolutional neural network (CNN) regression model to estimate PMA from local image patches extracted from the diffusion MRI dataset. These were combined to form a global prediction for each MRI scan. We found that CNN can reliably estimate PMA (Pearson’s r = 0.6, p < 0.05) from MRIs before 36 weeks of age (‘Early’ scans). These results revealed that the local fractional anisotropy (FA) measures of these very early scans preserved age specific information. Most interestingly, infants who were later diagnosed with CP were more likely to have an estimated younger brain age from ‘Early’ scans, the estimated age deviations were significantly different (Regression coefficient: −2.16, p < 0.05, corrected for actual age) compared to those infants who were not diagnosed with CP.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Infants born very preterm may be at high risk of structural and functional abnormalities of the brain, as well as adverse outcomes including Cerebral Palsy (CP) [1]. Currently, the diagnosis of CP or other motor and cognitive abnormalities are made at approximately 2 years of age [2]. Early detection of developmental abnormalities and thus earlier intervention and treatment are critical to improve outcomes for affected individuals. Brain imaging, such as magnetic resonance imaging (MRI), is one of the techniques to identify early markers of motor or cognitive outcome [3]. Finding an early biomarker specific to CP or other types of adverse outcomes is a challenging task [4]. Analyses of large datasets are needed to determine any overlooked and generalized features for early prediction of adverse neurodevelopmental outcome such as CP in preterm infants. For this task, cutting-edge techniques like machine learning or deep learning based technologies may play an important role.
Estimating morphological age and its deviation from the nominal gestational age by atlas based methods from brain MR images has proven useful for assessment of pathologies like lissencephaly [5]. Few studies that used CNN for estimating age (in years) directly from the MRI scans were on adult cohort [6, 7], however, these techniques have not been investigated in preterm infants or for prediction of infants at risk of CP. We propose to use a CNN trained with brain patches to estimate PMA of preterm born infants using brain diffusion MRI. The globally and locally estimated brain ages might be utilized to identify brain structures that indicate of PMA and to determine the correlation of local and global age deviations with clinical phenotype.
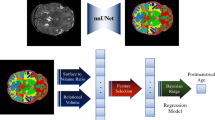
Here, we estimated the postmenstrual age (PMA) from diffusion MRI of preterm infants by a deep learning CNN based regression model. Infants were scanned at 2 time points, ‘Early’ (29.4–35.3 weeks) and ‘Term’ (age: 38.43–46.6 weeks) PMA. A CNN regression model was trained on a preterm cohort with no evidence of CP at 2 years corrected age, and then tested on the infants with or without a later diagnosis of CP separately. We compared our findings with a brain-volume based age estimation model. Overall, our aim was to investigate whether estimating PMA from MRI of preterm infants is possible in weekly resolution by a patch based CNN model and whether there is any difference in the range of estimated deviations between infants later diagnosed with CP compared to those who were not.
2 Method
2.1 MR Imaging Acquisition
Infants born <31 weeks gestational age (GA) were enrolled and scanned utilizing an MR compatible incubator equipped with a dedicated neonatal head coil (LMT Lammers Medical Technology, Lübeck, Germany) as part of a prospective cohort study [1]. Diffusion images were acquired, consisting of one low (b = 0 s/mm2) and 64 diffusion-weighted images (2000 s/mm2), in which the diffusion encoding gradients were uniformly distributed in space. Imaging parameters of the diffusion sequence were: field of view 224 × 224 mm, matrix 128 × 128, repetition time 9500 ms, echo time 130 ms and flip angle of 90°. Conventional MRI was conducted to assess brain abnormalities. A total of 119 infants underwent at least the ‘Early’ MRI. We excluded infants from our study who did not attend follow-up assessment at 2 years, or whose diffusion weighted images were of poor quality. Data of a total of 82 infants not diagnosed with CP (non-CP cohort) and 4 infants diagnosed with CP (CP cohort) at 2 years corrected age were analyzed.
Brain abnormalities were scored using conventional images [8], see Table 1. A histogram on the distribution of age is shown in Fig. 1.
2.2 MRI Preprocessing
MRI pre-processing procedures included removal of volumes affected by intra-volume motion, correction of between volume motion including rotation of the b-matrix, correction of image distortions due to susceptibility inhomogeneities using a field map, and detection and replacement of signal intensity outlier slices prior to resampling. Images were upsampled to 1.25 mm isotropic resolution and maps of fractional anisotropy (FA) were estimated using the diffusion tensor model. Brain masks were estimated from the non-diffusion-weighted images using registration to a study specific template and subsequent multi-atlas voting. Brain volumes were calculated from the brain masks. FA images were affinely registered to one of two study specific atlases created separately for the ‘Early’ and ‘Term’ time points. The same transformations were applied to the brain masks. All data were normalized to zero mean and unit variance.
2.3 Data Preprocessing for CNN and ‘Brain Volume’ Model
Our dataset for the non-CP cohort was composed of 142 scans (72 ‘Early’ and 70 ‘Term’ MRI). We used 80% of the scans for training, 10% for validation and 10% for test. Our CP cohort was small with 6 scans (2 ‘Early’ and 4 ‘Term’ MRI) from 4 infants. The same partitions were used for the brain-volume based linear regression model (‘Brain Volume’ model) for age estimation. For the CNN, 20 × 20 × 20 voxel non-overlapping patches were extracted from the scans within the brain using the brain masks, which were then used to train the network. The number of scans and patches for training, validation and test datasets are listed in Table 2.
2.4 Network Architecture
The CNN consisted of three 2D convolution layers with 3 × 3 kernel and three max-pooling layers with stride of 2. Dropout was used as regularization to prevent overfitting. ReLU activation was used for each of the convolutional layers. The final 3 layers were fully connected, which blended the parameters to combine the feature vectors. The output of the network was a scalar, which indicated the predicted brain age for each patch. The learning rate was 0.001 and ‘Adam’ optimizer was used. The details of this network architecture can be found in Table 3.
2.5 Implementation
The CNN was implemented in TensorFlow (1.5.0) on a clustered CPU computation environment. The end-to-end algorithm was written in Python. With CPU computation, the time for training was ~2 h and testing was ~2 s for each test dataset. For the ‘Brain Volume’ model, a scikit-learn based linear regression function was used.
2.6 Performance Measures
As a post-processing step (Fig. 2), a single estimated age value was assigned to each scan from the CNN estimation with maximum frequency (mode) over its patches. Pearson correlation coefficients were calculated between actual and predicted PMA for ‘Early’ and ‘Term’ patches and scans of ‘Validation’ and ‘Test’ datasets. Similarly, correlations were also measured for the ‘Brain Volume’ model. Bland-Altman plots were generated for analyzing residuals for both of the models. In addition, CNN prediction accuracies for patches and scans at different estimated deviation ranges were reported. Finally, brain age deviation for each scan was calculated from the difference between the model-predicted brain age and actual postmenstrual age for both of the models and compared between non-CP and CP group by histograms, fitted kernel density estimates (KDE) and general linear regression models.
3 Results
We first tested whether our CNN model can estimate the local and global PMA from MRI patches and scans respectively for both preterm infants with and without CP. CNN model predictions for patches for the non-CP ‘Validation’, non-CP ‘Test’ and CP scans are shown in Fig. 3A. The Pearson’s r between actual and predicted ages was (0.07, p = 0.1) for ‘Early’ non-CP ‘Test’ patches and (0.2, p < 0.05) for the ‘Term’ patches. This poor correlation can be attributed to a high variability in the predicted ages over local patches of any scan. Therefore, after combining the patch-based estimates into a single scan-based estimate (post-processing), r increased to 0.62 (p = 0.054) for the ‘Early’ non-CP ‘Test’ scans and 0.63 (p = 0.25) for ‘Term’ scans. When the ‘Test’ and ‘Validation’ datasets were combined, we found a significant correlation (r = 0.6, p < 0.05) for ‘Early’ scans and a poor correlation (r = 0.25 p = 0.3) for the ‘Term’ scans. Thus, despite the variability over local estimations, local patches in scans preserved age specific features in ‘Early’ MRI. The ‘Brain Volume’ model predictions also showed a strong correlation for both ‘Early’ (r = 0.75, p < 0.05) and ‘Term’ (r = 0.65, p = 0.23) non-CP ‘Test’ scans. For the combination of ‘Validation’ and ‘Test’ scans, r decreased to 0.61 (p < 0.05) for ‘Early’ scans and to 0.4 (p = 0.22) for ‘Term’ scans. Thus, both of the models showed a strong age predictability (in weeks) from ‘Early’ MRI. Predictions from ‘Term’ MRI were unreliable for the CNN model with the particular post-processing scheme and inaccurate for the ‘Brain Volume’ model as shown by their respective residual plots in Fig. 3D and E. Therefore, we excluded ‘Term’ predictions from further analysis.
A. Patch based predictions of PMA in non-CP (Validation and Test) and CP datasets. B and C. Scan based predictions (after post-processing) of PMA in non-CP and CP datasets by CNN and ‘Brain Volume’ model respectively. D & E: Residual plots for CNN and ‘Brain Volume’ model, respectively. F & G. Comparison of estimated age deviations between non-CP ‘Test’ and ‘CP’ group from CNN and ‘Brain Volume’ model respectively.
As a secondary analysis, we calculated the patch and scan based prediction accuracy for ‘Early’ scans only at different age deviations as shown in Tables 4 and 5. The age prediction accuracies for CNN model were 70% for non-CP (‘Test’) and 100% for CP scans within ±2w deviation. We then compared the distributions in estimated age deviations (for ‘Early’ scans only) between non-CP and CP scans for both of the models (Fig. 3F and G). Interestingly, for CNN model, ‘Early’ brain age was consistently underestimated for infants with CP, while it was either under- or overestimated for infants without CP (Fig. 3D and F). For ‘Brain Volume’ model, 50% of the ‘Early’ CP scans were underestimated (Fig. 3E and G). Thus, the CNN estimated age deviations for ‘Early’ scans were significantly reduced (Regression coefficient: −2.16, p < 0.05, corrected for actual age) in CP compared to non-CP (Test +Validation) while the ‘Brain Volume’ model estimated deviations were not (Regression coefficient = −1.58, p = 0.11, corrected for actual age). For the ‘Brain Volume’ model, the underestimation for CP scans seems to be related to a smaller brain volume than expected at an age, while CNN features related with underestimation are still to be explored as the MRI preprocessing steps essentially negate the effect of brain size. Nevertheless, these results represent that underestimated brain age preferably by CNN models might indicate risk of CP.
4 Discussion
We presented a CNN based postmenstrual age prediction approach for preterm infants and, to our knowledge, this is the first study to utilize the CNN estimations as a very early predictor of CP. The expected developmental abnormalities of the preterm brain with varying degrees of brain abnormalities made the age prediction task from local brain features challenging. Our study showed that both local brain feature based CNN regression model and total brain volume based linear regression model (‘Brain Volume’) reliably predicted PMA of the preterm infants from their ‘Early’ scans with strong correlations and reasonable prediction accuracy within ±2w deviation.
The Bland-Altman plots for CNN predictions (Fig. 3D) appear to show a systematic prediction error (overestimation for younger, underestimation for older) for ‘Early’ test scans but not for ‘Early’ validation scans. With only one train/validation/test split, it is difficult to determine whether this is coincidental. Cross-validations are required to verify these findings. More importantly, different types of post processing schemes, which biologically represent how the local age information in the brain is related to a global age, should be explored. Detailed studies on the patches, which are associated with the closest estimation of the actual age, should be conducted and thus only specific regions could be considered while predicting a global age. The brain age prediction was unreliable/inaccurate at term equivalent age for both of the models as indicated by lower Pearson’s r correlation and residuals (Fig. 3D and E). The more reliable prediction at younger age could be due to more rapid changes to the brain folding patterns or fractional anisotropy in the early period than later. As a consequence, there may not be enough information in the local features of the ‘Term’ scans for sufficiently reliable predictions; in addition, the expected abnormal developmental trajectory for the preterm might make the prediction harder. The ideal post-processing scheme might be different between ‘Early’ and ‘Term’ scans. In addition, the age band for ‘Term’ training was fairly narrow.
The most interesting finding is that while the non-CP dataset consisted of some patients with brain abnormality as shown in Table 1, the estimated age deviations from CNN models were significantly different between the ‘Early’ non-CP and CP groups of scans. The difference was not significant though for ‘Brain Volume’ model estimations. Nevertheless, the distributions from both of the models as shown in Fig. 3F and G revealed that CP cases are more likely to be underestimated than non-CP ones. In addition, it is noticeable that 100% of the CP scans (Early + Term) in CNN and ~70% of the CP scans in Brain Volume model were underestimated. This underestimation could be related to smaller brain volumes than normal, and for the CNN, any developmental delay in FA features. A number of previous studies [9, 10] reported the association of head circumference and developmental dysfunction related to CP, while with respect to FA features, previous data [11] showed that children with poor developmental outcomes at the age of 2 have lower FA in specific brain regions. Our study investigated these features at very early stages by utilizing local and global brain age deviations with an aim to find out a distinction line between non-CP and CP scans. It seems that underestimated brain age from the ‘Early’ scans preferably by CNN model could be one of the diagnostic features of CP, which in combination with other clinical scores could be indicative of CP disorder at a very early stage. Finally, it should be noted that all these observations were made on 2 ‘Early’ and 4 ‘Term’ CP scans and need to be verified on larger cohorts of preterm infants with CP.
One of the main advantages of a patch based CNN approach is that it might be able to identify the local brain patches that contain stronger features for age as well as the patches that show higher estimated deviations and thus could be potentially related to brain abnormalities. Future studies will explore the correlation of CNN model estimations and clinical findings in local brain regions. CNN training with larger samples and with multimodal MRI inputs will likely facilitate this kind of study.
References
George, J., et al.: PPREMO: a prospective cohort study of preterm infant brain structure and function to predict neurodevelopmental outcome. BMC Pediatr. 15(1), 123 (2015)
McIntyre, S., Morgan, C., Walker, K., Novak, I.: Cerebral Palsy-don’t delay. Dev. Disabil. Res. Rev. 17(2), 114–129 (2011)
George, J., et al.: Relationship between very early brain structure and neuromotor, neurological and neurobehavioral function in infants born <31 weeks gestational age. Early Hum. Dev. 117, 74–82 (2018)
Zhang, J.: Multivariate analysis and machine learning in Cerebral Palsy research. Front. Neurol. 8, 715 (2017)
Dittrich, E., et al.: A spatio-temporal latent atlas for semi-supervised learning of fetal brain segmentations and morphological age estimation. Med. Image Anal. 18(1), 9–21 (2014)
Cole, J., et al.: Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. NeuroImage 163, 115–124 (2017)
Huang, T., Chen, H., Fujimoto, R.: Age estimation from brain MRI images using deep learning. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia. IEEE (2017)
George, J., et al.: Validation of an MRI brain injury and growth scoring system in very preterm infants scanned at 29- to 35-week postmenstrual age. Am. J. Neuroradiol. 38(7), 1435–1442 (2017)
Jensen, A., Holmer, B.: White matter damage in 4,725 term-born infants is determined by head circumference at birth: the missing link. Obstet. Gynecol. Int. 2018, 1–12 (2018)
Kuban, K., et al.: Developmental correlates of head circumference at birth and two years in a cohort of extremely low gestational age newborns. J. Pediatr. 155(3), 344–349.e3 (2009)
Babcock, M., et al.: Injury to the preterm brain and cerebral palsy: clinical aspects, molecular mechanisms, unanswered questions, and future research directions. J. Child Neurol. 24(9), 1064–1084 (2009)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Crown
About this paper
Cite this paper
Saha, S. et al. (2018). Investigating Brain Age Deviation in Preterm Infants: A Deep Learning Approach. In: Melbourne, A., et al. Data Driven Treatment Response Assessment and Preterm, Perinatal, and Paediatric Image Analysis. PIPPI DATRA 2018 2018. Lecture Notes in Computer Science(), vol 11076. Springer, Cham. https://doi.org/10.1007/978-3-030-00807-9_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-00807-9_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00806-2
Online ISBN: 978-3-030-00807-9
eBook Packages: Computer ScienceComputer Science (R0)