Abstract
Treponema succinifaciens Cwyk and Canale-Parola 1981 is of interest because this strictly anaerobic, apathogenic member of the genus Treponema oxidizes carbohydrates and couples the Embden-Meyerhof pathway via activity of a pyruvate-formate lyase to the production of acetyl-coenzyme A and formate. This feature separates this species from most other anaerobic spirochetes. The genome of T. succinifaciens 6091T is only the second completed and published type strain genome from the genus Treponema in the family Spirochaetaceae. The 2,897,425 bp long genome with one plasmid harbors 2,723 protein-coding and 63 RNA genes and is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Similar content being viewed by others
Introduction
Strain 6091T (= DSM 2489 = ATCC 33096 = JCM 13475) is the type strain of Treponema succinifaciens [1,2]. Currently, there are 25 species placed in the genus Treponema [3]. The species epithet is derived from the Latin noun acidum succinicum meaning succinic acid and the Latin verb facio meaning to make, produce, referring to the succinic acid-producing property of the species [1]. T. succinifaciens was isolated from the colon of swine, and first described as small spirochete by Harris et al. in 1972 [4]. In 1974 it was published that strain 6091T belonged to a group of harmless inhabitants of the intestine of healthy pigs and had no pathogenic potential [5]. No further isolates have been described and strain 6091T was designated the type strain of the new species T. succinifaciens in 1979 [1]. Here we present a summary classification and a set of features for T. succinifaciens 6091T, together with the description of the complete genomic sequencing and annotation.
Classification and features
A representative genomic 16S rRNA sequence of T. succinifaciens was compared using NCBI BLAST [6] under default settings (e.g., considering only the high-scoring segment pairs (HSPs) from the best 250 hits) with the most recent release of the Greengenes database [7] and the relative frequencies of taxa and keywords (reduced to their stem [8]) were determined, weighted by BLAST scores. The most frequently occurring genera were Treponema (97.5%) and Spirochaeta (2.5%) (32 hits in total). Regarding the single hit to sequences from members of the species, the average identity within HSPs was 97.7%, whereas the average coverage by HSPs was 96.1%. Regarding the 14 hits to sequences from other members of the genus, the average identity within HSPs was 89.2%, whereas the average coverage by HSPs was 54.1%. Among all other species, the one yielding the highest score was Treponema socranskii (AY369254), which corresponded to an identity of 89.8% and an HSP coverage of 55.7%. (Note that the Greengenes database uses the INSDC (= EMBL/NCBI/DDBJ) annotation, which is not an authoritative source for nomenclature or classification.) The highest-scoring environmental sequence was EU462604 (‘Evolution mammals and their gut microbes Sumatran orangutan feces clone orang2 aai66a03’), which showed an identity of 99.6% and an HSP coverage of 91.6%. The most frequently occurring keywords within the labels of environmental samples which yielded hits were ‘gut’ (11.2%), ‘evolut, fece, mammal, microb’ (8.2%), ‘baboon, hamadrya’ (6.3%), ‘termit’ (5.1%) and ‘homogen’ (2.2%) (218 hits in total). The most frequently occurring keywords within the labels of environmental samples which yielded hits of a higher score than the highest scoring species were ‘gut’ (12.1%), ‘evolut, fece, mammal, microb’ (11.5%), ‘baboon, hamadrya’ (9.9%), ‘rumen’ (1.3%) and ‘termit’ (1.1%) (77 hits in total). These keywords fit to the ecological and physiological properties reported for strain 6091T in the original description [1].
Figure 1 shows the phylogenetic neighborhood of T. succinifaciens in a 16S rRNA based tree. The sequences of the four 16S rRNA gene copies in the genome differ from each other by up to seven nucleotides, and differ by up to 14 nucleotides from the previously published 16S rRNA sequence (M57738), which contains 26 ambiguous base calls.
Phylogenetic tree highlighting the position of T. succinifaciens relative to the type strains of the other species within the phylum ‘Spirochaetes’. The tree was inferred from 1,350 aligned characters [9,10] of the 16S rRNA gene sequence under the maximum likelihood (ML) criterion [11]. Rooting was done initially using the midpoint method [12] and then checked for its agreement with the current classification (Table 1). The branches are scaled in terms of the expected number of substitutions per site. Numbers adjacent to the branches are support values from 1,000 ML bootstrap replicates [13] (left) and from 1,000 maximum parsimony bootstrap replicates [14] (right) if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [15] are marked with one asterisk, those also listed as ‘Complete and Published’ (as well as the target genome) with two asterisks [16].
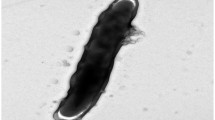
The cells of T. succinifaciens are of helical shape (0.3 × 4–8 µm) and usually exhibit irregular coiling (Figure 2). Cells are up to 16 µm long and also chains of cells may occur in culture [1]. T. succinifaciens is a Gram-negative and non spore-forming bacterium (Table 1). The organism displays temperature-dependent motility of translational, rotary and flexing movements; at 23–25°C no translational movement can be observed and rotation is slow, whereas at 37°C cells are very mobile (average velocity: 15µm/s) [1]. T. succinifaciens harbors two periplasmic fibrils inserted near each end of the cell [1]. The genome of T. succinifaciens contains 63 genes involved in motility (see below). The organism is a strictly anaerobic chemoorganotroph [1]. T. succinifaciens requires rumen fluid in media for good growth, replacement with a mixture of short-chain fatty acids leads to reduced growth yields [1]. The temperature range for growth is between 22°C and 43°C, with an optimum between 35°C and 39°C [1]. The organism is catalase-negative and does not grow in the presence of 6.5% NaCl [1]. T. succinifaciens requires CO2 for growth and is able to utilize arabinose, xylose, glucose, mannose, galactose, maltose, lactose, cellobiose, dextrin and starch for fermentation. Sugar alcohols, amino acids and other organic acids cannot be fermented by the organism [1]. Major fermentation products of glucose are acetate, formate, succinate and lactate, whereas pyruvate, acetoin and 2,3-butanediol are formed in minor amounts [1]. Assays of enzymatic activities showed that T. succinifaciens dissimilates glucose via the Embden-Meyerhof pathway [1]. It was shown that pyruvate is metabolized through the activity of pyruvate formate lyase to yield acetyl-coenzyme A and formate, which is in contrast to other spirochetes that degrade pyruvate to acetyl-coenzyme A, CO2 and H2 [1]. Furthermore, T. succinifaciens is capable of CO2 fixation for the production of succinate [1]. Also, the organism possesses enzymatic activity of adenine deaminase, phosphoribosyltransferase (for adenine, guanine and hypoxanthine), nucleotidase (for AMP, IMP and GMP), nucleoside phosphorylase (for adenosine, guanosine and inosine) and nucleoside hydrolase (for inosine and guanosine) [28]. Whether these activities are important for the survival of T. succinifaciens under nutrient deprivation or for adaptation to environmental stress is still unclear. An outer membrane-associated serine protease, which was found in several pathogenic spirochetes and also in T. succinifaciens, might be involved in the survival within the intestine, however, a role in pathogenesis can so far not be excluded [29]. T. succinifaciens is susceptible to penicillin G (4 units/ml), cephalotin (4 µg/ml) and chloramphenicol (4 µg/ml). Growth of the organism is not impaired by erythromycin (4 µg/ml), oxytetracycline (4 µg/ml), polymyxin B (40 units/ml), rifampin (4 µg/ml), streptomycin (4 µg/ml), tetracycline (4 µg/ml) or vancomycin (4 µg/ml) [1].
Chemotaxonomy
No chemotaxonomic information is currently available for T. succinifaciens.
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position [30], and is part of the Genomic Encyclopedia of Bacteria and Archaea project [31]. The genome project is deposited in the Genomes On Line Database [15] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Growth conditions and DNA isolation
T. succinifaciens strain 6091T, DSM 2489, was grown anaerobically in DSMZ medium 275 (Treponema succinifaciens medium) [32] at 37°C. DNA was isolated from 0.5–1 g of cell paste using MasterPure Gram-positive DNA purification kit (Epicentre MGP04100) following the standard protocol as recommended by the manufacturer with modification st/DL for cell lysis as described in Wu et al. 2009 [31]. DNA is available through the DNA Bank Network [33].
Genome sequencing and assembly
The genome was sequenced using a combination of Illumina and 454 sequencing platforms. All general aspects of library construction and sequencing can be found at the JGI website [34]. Pyrosequencing reads were assembled using the Newbler assembler (Roche). The initial Newbler assembly consisting of 134 contigs in two scaffolds was converted into a phrap assembly [35] by making fake reads from the consensus, to collect the read pairs in the 454 paired end library. Illumina sequencing data (3,531 Mb) was assembled with Velvet [36] and the consensus sequences were shredded into 1.5 kb overlapped fake reads and assembled together with the 454 data. The 454 draft assembly was based on 136.1 Mb 454 draft data and all of the 454 paired end data. Newbler parameters are -consed -a 50 -l 350 -g -m -ml 20. The Phred/Phrap/Consed software package [35] was used for sequence assembly and quality assessment in the subsequent finishing process. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected using gapResolution [34], Dupfinisher [37] or sequencing cloned bridging PCR fragments with subcloning. Gaps between contigs were closed by editing in Consed [35], by PCR and by Bubble PCR primer walks (J.-F. Chang, unpublished). A total of 305 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. Illumina reads were also used to correct potential base errors and increase consensus quality using a software Polisher developed at JGI [38]. The error rate of the completed genome sequence is less than 1 in 100,000. Together, the combination of the Illumina and 454 sequencing platforms provided 1,007.3 × coverage of the genome. The final assembly contained 486,725 pyrosequence and 36,577,056 Illumina reads.
Genome annotation
Genes were identified using Prodigal [39] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [40]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) non-redundant database, UniProt, TIGR-Fam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [41].
Genome properties
The genome consists of a 2,731,853 bp long chromosome and a 165.572 bp long plasmid both with a G+C content of 39.1% (Table 3, Figure 3 and Figure 4). Of the 2,786 genes predicted, 2,723 were protein-coding genes, and 63 RNAs; 115 pseudogenes were also identified. The majority of the protein-coding genes (57.8%) were assigned a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Graphical map of the chromosome (not drawn to scale with plasmid in Figure 4). From bottom to top: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Graphical map of the plasmid (not drawn to scale with chromosome in Figure 3). From bottom to top: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
References
Cwyk WM, Canale-Parola E. Treponema succinifaciens sp. nov., an anaerobic spirochete from the swine intestine. Arch Microbiol 1979; 122:231–239. PubMed doi:10.1007/BF00411285
Validation list No. 7. Int J Syst Bacteriol 1981; 31:382–383. doi:10.1099/00207713-31-3-382
Garrity G. NamesforLife. BrowserTool takes expertise out of the database and puts it right in the browser. Microbiol Today 2010; 37:9.
Harris DL, Kinyon JM, Mullin MT, Glock RD. Isolation and propagation of spirochetes from the colon of swine dysentery affected pigs. Can J Comp Med 1972; 36:74–76. PubMed
Harris DL, Kinyon JM. Significance of anaerobic spirochetes in the intestines of animals. Am J Clin Nutr 1974; 27:1297–1304. PubMed
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403–410. PubMed
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069–5072. PubMed doi:10.1128/AEM.03006-05
Porter MF. An algorithm for suffix stripping. Program: electronic library and information systems 1980; 14:130–137.
Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452–464. PubMed doi:10.1093/bioinformatics/18.3.452
Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540–552. PubMed
Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 2008; 57:758–771. PubMed doi:10.1080/10635150802429642
Hess PN, De Moraes Russo CA. An empirical test of the midpoint rooting method. Biol J Linn Soc Lond 2007; 92:669–674. doi:10.1111/j.1095-8312.2007.00864.x
Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184–200. doi:10.1007/978-3-642-02008-7_13
Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0 b10. Sinauer Associates, Sunderland, 2002.
Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346–D354. PubMed doi:10.1093/nar/gkp848
Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, Davidsen TM, DeBoy RT, Fouts DE, Haft DH, et al. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci USA 2004; 101:5646–5651. PubMed doi:10.1073/pnas.0307639101
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541–547. PubMed doi:10.1038/nbt1360
Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576–4579. PubMed doi:10.1073/pnas.87.12.4576
Garrity G, Holt JG. Phylum B17 Spirochaetes phy. nov. Garrity and Holt. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 138.
Ludwig W, Euzeby J, Whitman WG. Draft taxonomic outline of the Bacteroidetes, Planctomycetes, Chlamydiae, Spirochaetes, Fibrobacteres, Fusobacteria, Acidobacteria, Verrucomicrobia, Dictyoglomi, and Gemmatimonadetes. http://www.bergeys.org/outlines/Bergeys_Vol_4_Outline.pdf. Taxonomic Outline 2008.
Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225–420. doi:10.1099/00207713-30-1-225
Buchanan RE. Studies in the nomenclature and classification of bacteria. II. The primary subdivisions of the Schizomycetes. J Bacteriol 1917; 2:155–164. PubMed
Swellengrebel NH. Sur la cytologie comparée des spirochètes et des spirilles. Ann Inst Pasteur (Paris) 1907; 21:562–586.
Schaudinn F. Dtsch Med Wochenschr 1905; 31:1728. doi:10.1055/s-0029-1188418
Smibert RM. Genus III. Treponema Schaudinn 1905, 1728. In: Buchanan RE, Gibbons NE (eds), Bergey’s Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 175–184.
BAuA. 2005. Classification of bacteria and archaea in risk groups. TRBA 466 p. 349.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25–29. PubMed doi:10.1038/75556
Canale-Parola E, Kidder GW. Enzymatic activities for interconversion of purines in spirochetes. J Bacteriol 1982; 152:1105–1110. PubMed
Muniappa N, Duhamel GE. Outer membrane-associated serine protease of intestinal spirochetes. FEMS Microbiol Lett 1997; 154:159–164. PubMed doi:10.1111/J.1574-6968.1997.tb12638.x
Klenk HP, Göker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010; 33:175–182. PubMed doi:10.1016/j.syapm.2010.03.003
Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009; 462:1056–1060. PubMed doi:10.1038/nature08656
List of growth media used at DSMZ: http://www.dsmz.de/microorganisms/media_list.php
Gemeinholzer B, Dröge G, Zetzsche H, Haszprunar G, Klenk HP, Güntsch A, Berendsohn WG, Wägele JW. The DNA Bank Network: the start from a German initiative. Biopreservation and Biobanking 2011; 9:51–55. doi:10.1089/bio.2010.0029
The DOE Joint Genome Institute. http://www.jgi.doe.gov
Phrap and Phred for Windows, MacOS, Linux, and Unix. http://www.phrap.com
Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821–829. PubMed doi:10.1101/gr.074492.107
Han C, Chain P. 2006. Finishing repeat regions automatically with Dupfinisher. In: Proceeding of the 2006 international conference on bioinformatics & computational biology. Arabnia HR, Valafar H (eds), CSREA Press. June 26–29, 2006: 141–146.
Lapidus A, LaButti K, Foster B, Lowry S, Trong S, Goltsman E. POLISHER: An effective tool for using ultra short reads in microbial genome assembly and finishing. AGBT, Marco Island, FL, 2008.
Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119. PubMed doi:10.1186/1471-2105-11-119
Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455–457. PubMed doi:10.1038/nmeth.1457
Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271–2278. PubMed doi:10.1093/bioinformatics/btp393
Acknowledgements
We would like to gratefully acknowledge the help of Sabine Welnitz (DSMZ) for growing T. succinifaciens cultures. This work was performed under the auspices of the US Department of Energy Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, UT-Battelle and Oak Ridge National Laboratory under contract DE-AC05-00OR22725, as well as German Research Foundation (DFG) INST 599/1-2.
Author information
Authors and Affiliations
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Han, C., Gronow, S., Teshima, H. et al. Complete genome sequence of Treponema succinifaciens type strain (6091T). Stand in Genomic Sci 4, 361–370 (2011). https://doi.org/10.4056/sigs.1984594
Published:
Issue Date:
DOI: https://doi.org/10.4056/sigs.1984594