Abstract
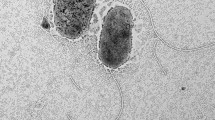
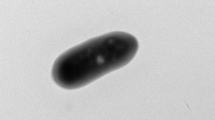
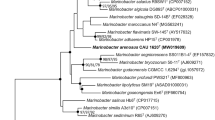
Marine sediment was collected from the Yellow Sea, Korea, and enriched using 1 mM thiosulfate under aerobic conditions. After 2 weeks, a facultative sulfur-oxidizing strain, BR13, was isolated. Phylogenetic analysis based on the 16S rRNA gene sequence showed that this strain belonged to the genus Marinobacter in the gamma-proteobacteria and was most closely related to Marinobacter pelagius H225T (99.5% 16S rRNA gene sequence similarity) and Marinobacter koreensis DD-M3T (97.7% 16S rRNA gene sequence similarity). The isolate was a sulfur-oxidizer, Gram-negative, aerobic, and slightly halophilic bacterium and grew in 0.5–9% NaCl at 7–37ºC. Strain BR13 shared some physiological and biochemical properties with Marinobacter pelagius H225T and Marinobacter koreensis DD-M3T. The G+C content of the genomic DNA of strain BR13 was 58.4 mol%. Strain BR13 possessed C16:0, summed feature 4 (C16:1 ω7c and/or C15:1iso 2OH), C16:0 N alcohol, and C18:1 ω9c as the major cellular fatty acids. Our results showed that strain BR13 (=KCTC 22253 =JCM 15135) is a novel facultative sulfur-oxidizing strain in the genus of Marinobacter.
Similar content being viewed by others
Abbreviations
- ASW:
-
artificial sea water
- JCM:
-
Japan Collection of Microorganisms
- KCTC:
-
Korean Collection for Type Cultures
- MA:
-
marine agar
- PCR:
-
polymerase chain reaction
References
Buck JD (1982) Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl Environ Microbiol 44, 992–993.
Campbell BJ, Engel AS, Porter ML, and Takai K (2006) The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol 4, 458–468.
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Friedrich CG, Rother D, Bardischewsky F, Quentmeier A, and Fischer J (2001) Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl Environ Microbiol 67, 2873–2882.
Gauthier MJ, Lafay B, Christen R, Fernandez L, Acquaviva M, Bonin P, and Bertrand JC (1992) Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int J Syst Bacteriol 42, 568–576.
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41, 95–98.
Handley KM, Héry M, and Lloyd JR (2009) Marinobacter santoriniensis sp. nov., an arsenate-respiring and arseniteoxidizing bacterium isolated from hydrothermal sediment. Int J Syst Evol Microbiol 59, 886–892.
Hiraishi A, Ueda Y, Ishihara J, and Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42, 457–469.
Kim BY, Weon HY, Yoo SH, Kim JS, Kwon SW, Stackebrandt E, and Go SJ (2006) Marinobacter koreensis sp. nov., isolated from sea sand in Korea. Int J Syst Evol Microbiol 56, 2653–2656.
Kimura M (1983) In The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK.
Kumar S, Tamura K, and Nei M (2004) MEGA3: Integrated Software for Molecular Evolutionary Genetics Analysis and Sequence Alignment. Brief Bioinform 5, 150–163.
Levring T (1946) Some culture experiments with Ulva and artificial seawater. Kungl Fysiografiska Sällsk Lund Förhandlingar 16, 45–56.
Mesbah M, Premachandran U, and Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39, 159–167.
MIDI (1999) In Sherlock Microbial Identification System Operating Manual (Version 3.0). pp. B8-B13. MIDI Inc., Newark, DE.
Park SJ, Kang CH, and Rhee SK (2006) Characterization of the microbial diversity in a Korean solar saltern by 16S rRNA gene analysis. J Microbiol Biotechnol 16, 1640–1645.
Ravenschlag K, Sahm K, Pernthaler J, and Amann R (1999) High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol 65, 3982–3989.
Saitou N and Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Bio Evol 4, 406–425.
Shieh WY, Jean WD, Lin YT, and Tseng M (2003) Marinobacter lutaoensis sp. nov., a thermotolerant marine bacterium isolated from a coastal hot spring in Lutao, Taiwan. Can J Microbiol 49 244–252.
Teske A, Brinkhoff T, Muyzer G, Moser DP, Rethmeier J, and Jannasch HW (2000) Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl Environ Microbiol 66, 3125–3133.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, and Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24, 4876–4882.
Voroteliak V, Cowley DM, and Florin TH (1993) Improved colorimetric determination of urinary thiosulfate to study intermediate sulfur metabolism in humans. Clin Chem 39, 2533–2534.
Weisburg WG, Barns SM, Pelletier DA, and Lane DJ (1991) 16S ribosomal DNA amplification for phylogenic study. J Bacteriol 173, 697–703.
Xu XW, Wu YH, Wang CS, Yang JY, Oren A, and Wu M (2008) Marinobacter pelagius sp. nov., a moderately halophilic bacterium, Int J Syst Evol Microbiol 58, 637–640.
Zhuang DC, Chen YG, Zhang YQ, Tang SK, Wu XL, Tan ZC, Li WJ, and Cui XL (2009) Marinobacter zhanjiangensis sp. nov., a marine bacterium isolated from sea water of a tidal flat of the South China Sea. Antonie Van Leeuwenhoek doi:10.1007/s10482-009-9344-5.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Choi, BR., Pham, V.H., Park, SJ. et al. Characterization of facultative sulfur-oxidizing Marinobacter sp. BR13 isolated from marine sediment of Yellow Sea, Korea. J. Korean Soc. Appl. Biol. Chem. 52, 309–314 (2009). https://doi.org/10.3839/jksabc.2009.055
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.3839/jksabc.2009.055