Abstract
Identification of subgroups of patients for whom treatment A is more effective than treatment B, and vice versa, is of key importance to the development of personalized medicine. Tree-based algorithms are helpful tools for the detection of such interactions, but none of the available algorithms allow for taking into account clustered or nested dataset structures, which are particularly common in psychological research. Therefore, we propose the generalized linear mixed-effects model tree (GLMM tree) algorithm, which allows for the detection of treatment-subgroup interactions, while accounting for the clustered structure of a dataset. The algorithm uses model-based recursive partitioning to detect treatment-subgroup interactions, and a GLMM to estimate the random-effects parameters. In a simulation study, GLMM trees show higher accuracy in recovering treatment-subgroup interactions, higher predictive accuracy, and lower type II error rates than linear-model-based recursive partitioning and mixed-effects regression trees. Also, GLMM trees show somewhat higher predictive accuracy than linear mixed-effects models with pre-specified interaction effects, on average. We illustrate the application of GLMM trees on an individual patient-level data meta-analysis on treatments for depression. We conclude that GLMM trees are a promising exploratory tool for the detection of treatment-subgroup interactions in clustered datasets.
Similar content being viewed by others
Introduction
In research on the efficacy of treatments for somatic and psychological disorders, the one-size-fits-all paradigm is slowly losing ground, and personalized or stratified medicine is becoming increasingly important. Stratified medicine presents the challenge of discovering which patients respond best to which treatments. This can be referred to as the detection of treatment-subgroup interactions (e.g., Doove, Dusseldorp, Van Deun, & Van Mechelen, 2014). Often, treatment-subgroup interactions are studied using linear models, such as factorial analysis of variance techniques, in which potential moderators have to be specified a priori, have to be checked one at a time, and continuous moderator variables have to be discretized. This may hamper identification of which treatment works best for whom, especially when there are no a priori hypotheses about treatment-subgroup interactions. As noted by Kraemer, Frank, and Kupfer (2006), there is a need for methods that generate instead of test such hypotheses.
Tree-based methods are such hypothesis-generating methods. Tree-based methods, also known as recursive partitioning methods, split observations repeatedly into groups so that they become increasingly similar with respect to the outcome within each group. Several tree-based methods take the mean of a continuous dependent variable or the majority class of a categorical dependent variable as the outcome, one of the earliest and most well-known examples being the classification and regression tree (CART) approach of Breiman, Friedman, Olshen, and Stone (1984). Other tree-based methods take the estimated parameters of a more complex model, of which the RECPAM (recursive partition and amalgamation) approach of Ciampi (1991) is the earliest example.
Due to the recursive nature of the splitting, the rectangular regions of a recursive partition can be graphically depicted as nodes in a decision tree, as shown in the artificial example in Fig. 1. The partition in Fig. 1 is rather simple, based on the values of two predictor variables: duration and anxiety. The resulting tree has a depth of two, as the longest path travels along two splits. Each of the splits in the tree is defined by a splitting variable and value. The first split in the tree separates the observations into two subgroups, based on the duration variable and a splitting value of eight, yielding two rectangular regions, represented by node 2 and node 5. Node 2 is an inner node, as the observations in this node are further split into terminal nodes 3 and 4, based on the anxiety variable. The observations in node 5 are not further split and this is therefore a terminal node.
If the partition in Fig. 1 would be used for prediction of a new observation, the new observation would be assigned to one of the terminal nodes according to its values on the splitting variables. The prediction is then based on the estimated distribution of the outcome variable within that terminal node. For example, the prediction may be the node-specific mean of a single continuous variable. In the current paper, we focus on trees where the terminal nodes consist of a linear (LM) or generalized linear model (GLM), in which case the predicted value for a new observation is determined by the node-specific parameter estimates of the (G)LM, while also adjusting for random effects.
Tree-based methods are particularly useful for exploratory purposes because they can handle many potential predictor variables at once and can automatically detect (higher-order) interactions between predictor variables (Strobl, Malley, & Tutz, 2009). As such, they are preeminently suited to the detection of treatment-subgroup interactions. Several tree-based algorithms for the detection of treatment-subgroup interactions have already been developed (Dusseldorp, Doove, & Van Mechelen, 2016; Dusseldorp & Meulman, 2004; Su, Tsai, Wang, Nickerson, & Li, 2009; Foster, Taylor, & Ruberg, 2011; Lipkovich, Dmitrienko, Denne, & Enas, 2011; Zeileis, Hothorn, & Hornik, 2008; Seibold, Zeileis, & Hothorn, 2016; Athey & Imbens 2016). Also, Zhang, Tsiatis, Laber, and Davidian (2012b) and Zhang, Tsiatis, Davidian, Zhang, and Laber (2012a) have developed a flexible classification-based approach, allowing users to select from a range of statistical methods, including trees.
In many instances, researchers may want to detect treatment-subgroup interactions in clustered or nested datasets, for example in individual-level patient data meta-analyses, where datasets of multiple clinical trials on the same treatments are pooled. In such analyses, the nested or clustered structure of the dataset should be taken into account by including study-specific random effects in the model, prompting the need for a mixed-effects model (e.g., Cooper & Patall 2009; Higgins, Whitehead, Turner, Omar, & Thompson, 2001). In linear models, ignoring the clustered structure may lead, for example, to biased inference due to underestimated standard errors (e.g., Bryk & Raudenbush, 1992). For tree-based methods, ignoring the clustered structure has been found to result in the detection of spurious subgroups and inaccurate predictor variable selection (e.g., Sela & Simonoff, 2012; Martin, 2015). However, none of the purely tree-based methods for treatment-subgroup interaction detection allow for taking into account the clustered structure of a dataset. Therefore, in the current paper, we present a tree-based algorithm that can be used for the detection of interactions and non-linearities in GLMM-type models: generalized linear mixed-effects model trees, or GLMM trees.
The GLMM tree algorithm builds on model-based recursive partitioning (MOB, Zeileis et al., 2008), which offers a flexible framework for subgroup detection. For example, GLM-based MOB has been applied to detect treatment-subgroup interactions for the treatment of depression (Driessen et al., 2016) and amyotrophic lateral sclerosis (Seibold et al., 2016). In contrast to other purely tree-based methods (e.g., Zeileis et al., 2008; Su et al., 2009; Dusseldorp et al., 2016), GLMM trees allow for taking into account the clustered structure of datasets. In contrast to previously suggested regression trees with random effects (e.g., Hajjem, Bellavance, & Larocque, 2011; Sela & Simonoff, 2012), GLMM trees allow for treatment effect estimation, with continuous as well as non-continuous response variables.
The remainder of this paper is structured into four sections: In the first section, we introduce the GLMM tree algorithm using an artificial motivating dataset with treatment-subgroup interactions. In the second section, we compare the performance of GLMM trees with that of three other methods: MOB trees without random effects, mixed-effects regression trees (MERTs) and linear mixed-effects models with pre-specified interactions. In the third section, we apply the GLMM tree algorithm to an existing dataset of a patient-level meta-analysis on the effects of psycho- and pharmacotherapy for depression. In the fourth and last section, we summarize the results and discuss limitations and directions for future research. In the Appendix, we provide a glossary explaining abbreviations and mathematical notation used in the current paper. Finally, a tutorial on how to fit GLMM trees using the R package glmertree is included as supplementary material. In the tutorial, the artificial motivating dataset is used, allowing users to recreate the trees and models to be fitted in the next section.
GLMM tree algorithm
Artificial motivating dataset
We will use an artificial motivating dataset with treatment-subgroup interactions to introduce the GLMM tree algorithm. This dataset consists of a set of observations on N = 150 patients, who were randomly assigned to one of two treatment alternatives (Treatment 1 or Treatment 2). The treatment outcome is represented by the variable depression, quantifying post-treatment depressive symptomatology. The potential moderator variables are duration, age, and anxiety. Duration reflects the number of months the patient has been suffering from depression prior to treatment, age reflects patients’ age in years at the start of treatment, and anxiety reflects patients’ total scores on an anxiety inventory administered before treatment. Summary statistics of these variables are provided in Table 1. Each patient was part of one of ten clusters, each having a different value for the random intercept, which were generated from a standard normal distribution and uncorrelated with the partitioning variables.
The outcome variable was generated such that there are three subgroups with differential treatment effectiveness, corresponding to the terminal nodes in Fig. 1: For the first subgroup of patients (node 3) with short duration (≤ 8) and low anxiety scores (≤ 10), Treatment 1 leads to lower post-treatment depression than in Treatment 2 (true mean difference = 2). For the second subgroup of patients (node 4) with short duration but high anxiety scores (> 10), post-treatment depression is about equal in both treatment conditions (true mean difference = 0). For the third subgroup of patients (node 5) with long duration (> 8 months), Treatment 2 leads to lower post-treatment depression than Treatment 1 (true mean difference = −2.5). Thus, duration and anxiety are true partitioning or moderator variables, whereas age is not. Anticipating the final results of our analyses, the treatment-subgroup interactions are depicted in Fig. 4, which shows the GLMM tree that accurately recovered the treatment-subgroup interactions.
Model-based recursive partitioning
The rationale behind MOB is that a single global GLM (or other parametric model) may not describe the data well, and when additional covariates are available it may be possible to partition the dataset with respect to these covariates, and find better-fitting models in each cell of the partition. For example, to assess the effect of treatment, we may first fit a global GLM where the treatment indicator has the same effect/coefficient on the outcome for all observations. Subsequently, the data may be partitioned recursively with respect to other covariates, leading to separate models with different treatment effects/coefficients in each subsample.
More formally, in a single global GLM, the expectation μ i of outcome y i given the treatment regressor x i is modeled through a linear predictor and suitable link function:
where x i⊤β is the linear predictor for observation i and g is the link function. β is a vector of fixed-effects regression coefficients. For simplicity, in the current paper we focus on two treatment groups and no further covariates in the GLM, so that in our illustrations x i and β both have length 2. For the continuous response variable in the motivating data set, we employ the identity link function and assume a normal distribution for the error (denoted by 𝜖 i = y i − μ i ) with mean zero and variance σ 𝜖2. Thus, the first element of β then corresponds to the mean of the linear predictor in the first treatment group and the second element corresponds to the mean difference in the linear predictor between the first and second treatment groups. However, the model can easily accommodate additional treatment conditions and covariates, as well as binary or count/Poisson outcome variables.
Obviously, such a simple, global GLM will not fit the data well, especially in the presence of moderators. For expository purposes, however, we take it as a starting point to illustrate MOB. The global GLM fitted to the motivating example dataset is depicted in Fig. 2. As the boxplots show, there is little difference between the global effects of the two treatments and there is considerable residual variance.
Example of a globally estimated GLM based on the artificial motivating dataset (N = 150). The x-axis represents treatment group, the y-axis represents treatment outcome (post-treatment depression). The dot for Treatment 1 represents the first element of the fixed-effects coefficient vector β, the slope of the regression line represents the second element of β
The MOB algorithm can be used to partition the dataset using additional covariates and find better-fitting local models. To this end, the MOB algorithm tests for parameter stability with respect to each of a set of auxiliary covariates, also called partitioning variables, which we will denote by U. When the partitioning is based on a GLM, instabilities are differences in \(\hat {\beta }\) across partitions of the dataset, which are defined by one or more auxiliary covariates not included in the linear predictor. To find these partitions, the MOB algorithm cycles iteratively through the following steps (Zeileis et al., 2008): (1) fit the parametric model to the dataset, (2) statistically test for parameter instability with respect to each of a set of partitioning variables, (3) if there is some overall parameter instability, split the dataset with respect to the variable associated with the highest instability, (4) repeat the procedure in each of the resulting subgroups.
In step (2) a test statistic quantifying parameter instability is calculated for every potential partitioning variable. As the distribution of these test statistics under the null hypothesis of parameter stability is known, a p value for every partitioning variable can be calculated. Note that a more in-depth discussion of the parameter stability tests is beyond the scope of this paper, but can be found in Zeileis and Hornik (2007) and Zeileis et al. (2008).
If at least one of the partitioning variables yields a p value below the pre-specified significance level α, the dataset is partitioned into two subsets in step (3). This partition is created using \(U_{k^{*}}\), the partitioning variable with the minimal p value in step (2). The split point for \(U_{k^{*}}\) is selected by taking the value that minimizes the instability as measured by the sum of the values of two loss functions, one for each of the resulting subgroups. In other words, the loss function is minimized separately in the two subgroups resulting from every possible split point and the split point yielding the minimum sum of the loss functions is selected. In step (4), steps (1) through (3) are repeated in each partition, until the null hypothesis of parameter stability can no longer be rejected (or the subsets become too small).
The partition resulting from application of MOB can be depicted as a decision tree. If the partitioning is based on a GLM, the result is a GLM tree, with a local fixed-effects regression model in every j-th (j = 1,…,J) terminal node:
To illustrate, we fitted a GLM tree on the artificial motivating dataset. In addition to the treatment indicator and treatment outcome used to fit the earlier GLM, we specified the anxiety, duration and age variables as potential partitioning variables. Figure 3 shows the resulting GLM tree. MOB partitioned the observations into four subgroups, each with a different estimate β j . Age was correctly not identified as a partitioning variable and the left- and rightmost nodes are in accordance with the true treatment-subgroup interactions described above. However, the two nodes in the middle represent an unnecessary split and thus do not represent true subgroups, possibly due to the dependence of observations within clusters not being taken into account.
GLM tree grown on the artificial motivating dataset. The x-axes in the terminal nodes represent the treatment group, the y-axes represent the treatment outcome (post-treatment depression). Three additional covariates (pre-treatment anxiety, duration, and age) were used as potential splitting variables, of which two (duration and age) were selected
Including random effects
For datasets containing observations from multiple clusters (e.g., trials or research centers), application of a mixed-effects model would be more appropriate. The GLM in Eq. 2 is then extended to include cluster-specific, or random effects:
For a random-intercept only model, z i is a unit vector of length M, of which the m-th element takes a value of 1, and all other elements take a value of 0; m (m = 1,…,M) denotes the cluster which observation i is part of. Further, b is a random vector of length M, each m-th element corresponding to the random intercept for cluster m. For simplicity, we employ a cluster-specific intercept only, but further random effects can easily be included in z i . Furthermore, within the GLMM it is assumed that b is normally distributed, with mean zero and variance σ b2 and that the errors 𝜖 have constant variance across clusters. The parameters of the GLMM can be estimated with, for example, maximum likelihood (ML) and restricted ML (REML).
Although the random-effects part of the GLMM in Eq. 4 accounts for the nested structure of the dataset, the global fixed-effects part x i⊤β may not describe the data well. Therefore, we propose the GLMM tree model, in which the fixed-effects part may be partitioned as in Eq. 3 while still adjusting for random effects:
In the GLMM tree model, the fixed effects β j are local parameters, their value depending on terminal node j, but the random effects b are global. To estimate the parameters of this model, we take an approach similar to that of the mixed-effects regression tree (MERT) approach of Hajjem et al. (2011) and Sela and Simonoff (2012). In the MERT approach, the fixed-effects part of a GLMM is replaced by a CART tree with constant fits in the nodes, and the random-effects parameters are estimated as usual. To estimate a MERT, an iterative approach is taken, alternating between (1) assuming random effects known, allowing for estimation of the CART tree, and (2) assuming the CART tree known, allowing for estimation of the random-effects parameters.
For estimating GLMM trees, we take this approach two steps further: (1) Instead of a CART tree with constant fits to estimate the fixed-effects part of the GLMM, we use a GLM tree. This allows not only for detection of differences in intercepts across terminal nodes but also for detection of differences in slopes such as treatment effects. (2) By using generalized linear (mixed) models, the response may also be a binary or count variable instead of a continuous variable. The GLMM tree algorithm takes the following steps to estimate the model in Eq. 5:
- Step 0::
-
Initialize by setting r and all values \(\hat {b}_{(r)}\) to 0.
- Step 1::
-
Set r = r + 1. Estimate a GLM tree using \(z_{i}^{\top }\hat {b}_{(r-1)}\) as an offset.
- Step 2::
-
Fit the mixed-effects model g(μ i j ) = x i⊤β j + z i⊤b with terminal node j(r) from the GLM tree estimated in Step 1. Extract posterior predictions \(\hat {b}_{(r)}\) from the estimated model.
- Step 3::
-
Repeat Steps 1 and 2 until convergence.
The algorithm initializes by setting b to 0, since the random effects are initially unknown. In every iteration, the GLM tree is re-estimated in step (1) and the fixed- and random-effects parameters are re-estimated in step (2). Note that the random effects are not partitioned, but estimated globally. Only the fixed effects are estimated locally, within the cells of the partition. Convergence of the algorithm is monitored by computing the log-likelihood criterion of the mixed-effects model in Eq. 5. Typically, this converges if the tree does not change from one iteration to the next.
In Fig. 4, the result of applying the GLMM tree algorithm to the motivating dataset is presented. In addition to the treatment indicator, treatment outcome and the potential partitioning variables, the GLMM tree algorithm has also taken a random intercept with respect to the cluster indicator into account. As a result, the dependence between observations is taken into account, the true treatment subgroups have been recovered and the spurious split involving the anxiety variable no longer appears in the tree.
GLMM tree of the motivating example dataset. The x-axes represent the treatment group, the y-axes represent the treatment outcome (post-treatment depression). Three covariates (anxiety, duration and age) were used as potential splitting variables, and the clustering structure was taken into account by estimating random intercepts
Simulation-based evaluation
To assess the performance of GLMM trees, we carried out three simulation studies: In Study I we assessed and compared the accuracy of GLMM trees, linear-model based MOB (LM trees) and mixed-effects regression trees (MERTs) in datasets with treatment-subgroup interactions. In Study II, we assessed and compared the type I error of GLMM trees and linear-model based MOB in datasets without treatment-subgroup interactions. In Study III, we assessed and compared the performance of GLMM trees and linear mixed-effects models (LMMs) with pre-specified interactions in datasets with piecewise and continuous interactions. As the outcome variable was continuous in all simulated datasets, the GLMM tree algorithm and trees resulting from its application will be referred to as LMM tree(s).
General simulation design
In all simulation studies, the following data-generating parameters were varied:
-
1.
Sample size: N = 200, N = 500, N = 1000.
-
2.
Number of potential partitioning covariates U 1 through U K : K = 5 and K = 15.
-
3.
Intercorrelation between the potential partitioning covariates U 1 through U K : \(\rho _{U_{k},U_{k^{\prime }}}=0.0\), \(\rho _{U_{k},U_{k^{\prime }}}=0.3\).
-
4.
Number of clusters: M = 5, M = 10, M = 25.
-
5.
Population standard deviation (SD) of the normal distribution from which the cluster-specific intercepts were drawn: σ b = 0, σ b = 5, σ b = 10.
-
6.
Intercorrelation between b and one of the U k variables: b and all U k covariates uncorrelated, b correlated with one of the U k covariates (r = .42).
Following the approach of Dusseldorp and Van Mechelen (2014), all partitioning covariates U 1 through U K were drawn from a multivariate normal distribution with means μ U 1 = 10, μ U 2 = 30, μ U 4 = −40, and μ U 5 = 70. The means of the other potential partitioning covariates (U 3 and, depending on the value of K, also U 6 through U 15) were drawn from a discrete uniform distribution on the interval [−70,70]. All covariates U 1 through U 15 had the same standard deviation: σ U k = 10.
To generate the cluster-specific intercepts, we partitioned the sample into M equally sized clusters, conditional on one of the variables U 1 through U 5, producing the correlations in the sixth facet of the simulation design. For each cluster, a single value b m was drawn from a normal distribution with mean 0 and the value of σ b given by the fifth facet of the simulation design. If b was correlated with one of the potential partitioning variables, the correlated variable was randomly selected.
For every observation, we generated a binomial variable (with probability 0.5) as an indicator for treatment type. Random errors 𝜖 were drawn from a normal distribution with μ 𝜖 = 0 and σ 𝜖 = 5. The value of the outcome variable y i was calculated as the sum of the random intercept, (node-specific) fixed effects and the random error term.
Due to the large number of cells in the simulation design, the most important predictors of accuracy were determined by means of ANOVAs and/or GLMs. The most important predictors of accuracy where then assessed through graphical displays. The ANOVAs and GLMs included main effects of algorithm type and the parameters of the data-generating process, as well as first-order interactions between algorithm type and each of the data-generating parameters.
Software
R (R Core Team, 2016) was used for data generation and analyses. The partykit package (version 1.0-2; Hothorn & Zeileis, 2015, 2016) was employed for estimating LM trees, using the lmtree function. For estimation of LMM trees, the lmertree function of the glmertree package (version 0.1-0; Fokkema & Zeileis, 2016; available from R-Forge) was used. The significance level α for the parameter instability tests was set to 0.05 for all trees, with a Bonferroni correction applied for multiple testing. The latter adjusts the p values of the parameter stability tests by multiplying these by the number of potential partitioning variables. The minimum number of observations per node in trees was set to 20 and maximum tree depth was set to three, thus limiting the number of terminal nodes to eight in every tree.
The REEMtree package (version 0.9.3; Sela & Simonoff, 2011) was employed for estimating MERTs, using default settings. For estimating LMMs, the lmer function from the lme4 package (version 1.1-7; Bates, Mächler, Bolker, & Walker, 2015; Bates et at., 2017) was employed, using restricted maximum likelihood (REML) estimation. The lmerTest package (version 2.0-32; Kuznetsova, Brockhoff, & Christensen, 2016) was used to assess statistical significance of fixed-effects predictors in LMMs in Study III. The lmerTest package calculates effective degrees of freedom and p values based on Satterthwaite approximations.
Study I: Performance of LMM trees, LM trees, and MERTs in datasets with treatment-subgroup interactions
Method
Treatment-subgroup interaction design
For generating datasets with treatment-subgroup interactions, we used a design from Dusseldorp and Van Mechelen (2014), which is depicted in Fig. 5. Figure 5 shows four terminal subgroups, characterized by values of the partitioning variables U 2, and U 1 or U 5. Two of the subgroups have mean differences in treatment outcome, indicated by a non-zero value of β j1, and two subgroups do not have mean differences in treatment outcome, indicated by a β j1 value of 0.
Data-generating model for treatment-subgroup interactions. U 1, U 2, and U 5 represent the (true) partitioning variables; β represents the (true) fixed effects, with β j0 representing the intercept and β j1 representing the slope; parameter d j denotes the node-specific standardized mean difference between the outcomes of Treatment 1 and 2 (i.e., β j1/σ 𝜖 )
In this simulation design, some of the potential partitioning covariates are true partitioning covariates, the others are noise variables. Therefore, in addition to the General simulation design, the following facet was added in this study:
-
6.
Intercorrelation between b and one of the U k variables: b and all U k covariates uncorrelated, b correlated with one of the true partitioning covariates (U 1, U 2 or U 5), b correlated with one of the noise variables (U 3 or U 4).
To assess the effect of the magnitude of treatment-effect differences, the following facet was added in this study:
-
7.
Two levels for the mean difference in treatment outcomes: The absolute value of the treatment-effect difference was varied to be |β j1| = 2.5 (corresponding to a medium effect size, Cohen’s d = 0.5; Cohen, 1992) and |β j1| = 5.0 (corresponding to a large effect size; Cohen’s d = 1.0).
For each cell of the design, 50 datasets were generated. In every dataset, the outcome variable was calculated as y i = x i⊤β j + z i⊤b m + 𝜖 i .
Assessment of performance
Performance of the algorithms was assessed by means of tree size, tree accuracy, and predictive accuracy. An accurately recovered tree was defined as a tree with (1) seven nodes in total, (2) the first split involving variable U 2 with a value of 30 ± 5, (3) the next split on the left involving variable U 1 with a value of 17 ± 5, and (4) the next split on the right involving variable U 5 with a value of 63 ± 5. The allowance of ± 5 equals plus or minus half the population SD of the partitioning variable (\(\sigma _{U_{k}}\)).
For MERT, the number of nodes and tree accuracy was not assessed, as the treatment-subgroup interaction design in Fig. 5 corresponds to a large number of regression tree structures, that would all be different but also correct. Therefore, performance of MERTs was only assessed in terms of predictive accuracy.
Predictive accuracy of each method was assessed by calculating the correlation between true and predicted treatment-effect differences. To prevent overly optimistic estimates of predictive accuracy, predictive accuracy was assessed using test datasets. Test datasets were generated from the same population as training datasets, but test observations were not drawn from the same clusters as the training observations, but from ‘new’ clusters.
The best approach for including treatment effects in MERTs is not completely obvious. Firstly, a single MERT may be fitted, where treatment is included as one of the potential partitioning variables. Predictions of treatment-effect differences can then be obtained by dropping test observations down the tree twice, once for every level of the treatment indicator. Secondly, two MERTs may be fitted: one using observations in the first treatment condition and one using observations in the second treatment condition. Predictions of treatment-effect differences can then be obtained by dropping a test observation down each of the two trees. We tried both approaches: the second approach yielded higher predictive accuracy, as the first approach often did not pick up the treatment indicator as a predictor. Therefore, we have taken the second approach of fitting two MERTs to each dataset in our simulations.
Results
Tree size
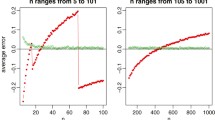
The average size of LMM trees was 7.15 nodes (SD = 0.61), whereas the average size of LM trees was 8.15 nodes (SD = 2.05), indicating that LM trees tend to involve more spurious splits than LMM trees. The effects of the most important predictors of tree size are depicted in Fig. 6. The average size of LMM trees was close to the true tree size in all conditions. In the absence of random effects, this was also the case for LM trees. In the presence of random effects that are correlated to a (potential) partitioning variable, LM trees start to create spurious splits, especially with larger σ b values. In the presence of random effects that are uncorrelated to the other variables in the model, LM trees lack power to detect treatment-subgroup interactions if sample size is small (i.e., N = 200). With larger sample sizes, LM trees showed about the true tree size, on average. Tree size of MERTs was not assessed, as a single true tree size for MERTs could not be derived from the design in Fig. 5.
Average tree size of LM and LMM trees. The y-axes represent the average number of nodes in trees; x-axes represent the population standard deviation of the normal distribution from which the cluster-specific intercepts were drawn; rows represent the correlation between the random intercepts and one of the partitioning variables; columns represent sample size. The horizontal line in each graph indicates the ’true’ tree size (total number of nodes in the tree used for generating the interactions)
Accuracy of recovered trees
The estimated probability that a dataset was erroneously not partitioned (type II error) was 0 for both algorithms. For the first split, LMM trees selected the true partitioning variable (U 2) in all datasets, and LM trees in all but one datasets. The mean splitting value of the first split was 29.94 for LM as well as LMM trees, which is very close to the true splitting value of 30 (Fig. 5).
Further splits were more accurately recovered by LMM trees yielding 90.40% accuracy for the full partition comparted to only 61.44% for LM trees. The effects of the four most important predictors of tree accuracy are depicted in Fig. 7. In the absence of random effects, LM and LMM trees were about equally accurate. In the presence of random effects, LM trees were much less accurate than LMM trees when random effects were correlated with a partitioning covariate. When random intercepts were not correlated with one of the U k variables, LMM trees outperformed LM trees only when sample size was small (i.e., N = 200). Tree accuracy of MERTs was not assessed, as a single accurate tree structure for MERTs could not be derived from the design in Fig. 5.
Tree accuracy of LM and LMM trees in simulated datasets. The y-axes represent the proportion of datasets in which the true tree was accurately recovered; x-axes represent the population standard deviation of the normal distribution from which the cluster-specific intercepts were drawn; rows represent dependence between the random intercepts (b) and one of the partitioning variables U k ; columns represent sample size
Predictive accuracy
The predicted treatment-effect differences of LMM trees show an average correlation of 0.93 (SD = .13) with the true differences. LM trees and MERTs show lower accuracy, with an average correlations of 0.88 (SD = .19) and 0.75 (SD = .21), respectively. The most important predictors of predictive accuracy are depicted in Fig. 8. Performance of all three algorithms improves with increasing sample size and treatment-effect differences. Furthermore, LMM trees and MERTs are not much affected by the presence and magnitude of random effects in the data. LMM trees perform most accurately in most conditions and are never outperformed by the other methods. MERTs perform the least accurate in most conditions and never outperform the other methods, but the differences in accuracy become less pronounced with larger sample and effect sizes.
Average predictive accuracy of MERT, LM and LMM trees in simulated datasets. The y-axes represent the average correlation between the true and predicted treatment-effect differences; x-axes represent the population standard deviation of the normal distribution from which the cluster-specific intercepts were drawn; rows represent absolute treatment-effect differences in subgroups with treatment-effect differences; columns represent sample size
Study II: Type I error of LM and LMM trees
Method
Design
In the second simulation study, we assessed the type I error rate of LM and LMM tree. In the datasets in this study, there was only a main effect of treatment in the population. Put differently, there was only a single global value of β j = β in every dataset. A type I error was defined as the proportion of datasets without treatment-subgroup interactions which were erroneously partitioned by the algorithm.
To assess the effect of the treatment-effect difference β, in addition to the General simulation design, the following facet was added in this study:
-
7.
Two levels for β, the global mean difference in treatment outcomes: β = 2.5 (corresponding to a medium effect size, Cohen’s d = 0.5) and β = 5.0 (corresponding to a large effect size; Cohen’s d = 1.0).
For each cell in the simulation design, 50 datasets were generated. In every dataset, the outcome variable was calculated as y i = x i⊤β + z i⊤b m + 𝜖 i .
Assessment of performance
To assess the type I error rates of LM and LMM trees, tree sizes were calculated and trees of size > 1 were classified as type I errors. The nominal type I error rate for both LM and LMM trees equals 0.05, corresponding to the pre-specified significance level α for the parameter instability tests.
Results
In datasets without treatment-subgroup interactions, average tree size was 1.09 (SD = 0.44) for LMM trees, and 2.02 (SD = 1.68) for LM trees. The average type I error rate was only 0.04 for LMM trees, and 0.33 for LM trees. Main predictors of type I error are depicted in Fig. 9, which shows that LMM trees have a type I error rate somewhat below the pre-specified α level in all conditions. The same goes for LM trees, when random effects are absent, or uncorrelated to one of the partitioning covariates. When the random intercept is correlated with one of the potential partitioning covariates, the type I error rapidly increases for LM trees. With increasing sample size or random-effects variance, LM trees will yield a larger number of spurious splits.
Type-I error rate of LM and LMM trees in simulated datasets. The y-axes represent the average type I error (or false-positive) rate, x-axes represent the population standard deviation of the normal distribution from which the cluster-specific intercepts were drawn represent; rows represent dependence between random intercepts (b) and one of the partitioning variables U k ; columns represent sample size
Study III: Recovery of piecewise and continuous interactions by LMM trees and LMMs with pre-specified interactions
Method
Interaction design
The interactions in Study I (Fig. 5) can be referred to as piecewise interactions, as their effect is a stepwise function of the moderator (partitioning) variables. Trees are preeminently suited for recovering such piecewise or subgroup interactions, but may have difficulty when the true interactions are continuous functions of moderator variables (for example, U 1 ⋅ U 2). At the same time, linear regression models with pre-specified interaction terms may perform well in recovering continuous interactions, but may have difficulty in recovering piecewise interactions. Therefore, in the third simulation study, in addition to the General simulation design described above, the following facet was added:
-
7.
Three levels for interaction type: continuous, piecewise and combined piecewise-continuous interactions.
To generate datasets with purely piecewise interactions, the same partition as in Study I (Fig. 5) was used. In other words, the outcome variable in this design was calculated as y i = x i⊤β j + z i⊤b + 𝜖 i , with the value of β j depending on the values of U 2, U 1 and U 5.
For generating datasets with both piecewise and continuous interactions, the partition as depicted in Fig. 5 was also used. In addition, the fixed-effects part x i⊤β j in each of the terminal nodes now comprised continuous main and (treatment) interaction effects of the partitioning variables. In other words, the partitioning variables U 2, U 1 and U 5 also appear in the linear predictor x i , as part of the terms presented in Table 2. The corresponding node-specific β j parameters are also presented in Table 2. The β j values were chosen to yield the same treatment-subgroup means as in Fig. 5. The interaction terms were created using centered U k variables, calculated by subtracting their variable means. Again, the outcome variable was calculated as y i = x i⊤β j + z i⊤b + 𝜖 i .
In datasets with purely continuous interactions, β has a global value and no subscript, comprising only purely continuous main and interaction effects, as shown by terms and the single column for β in Table 2. The outcome variable was calculated as y i = x i⊤β + z i⊤b + 𝜖 i .
Furthermore, in this simulation study, the number of cells in the design was reduced by limiting the fourth facet of the data-generating design to a single level (M = 25 clusters), as Study I and II indicated no effects of the number of clusters. The fifth facet of the data-generating design was limited to two levels (σ b = 2.5 and σ b = 7.5). For every cell of the design, 50 datasets were generated.
LMMs with pre-specified interactions
LMMs were estimated by specifying main effects for all covariates U k and the treatment indicator, first-order interactions between all pairs of covariates U k , and second-order interactions between all pairs of covariates U k and treatment. Continuous predictor variables were centered by subtracting the mean value, before calculating and including the interaction term in the LMM.
Assessment of performance
Predictive accuracy was assessed in terms of the correlation between the true and predicted treatment-effect differences in test datasets. As full LMMs may be likely to overfit, LMMs were refitted on the training data, using only the predictors with p values < 0.05 in the original LMM. Predictions for test observations were obtained using the refitted LMMs.
Results
On average, LMM trees showed somewhat higher accuracy: the average correlation between true and predicted treatment-effect differences was 0.54 (SD = .40) for LMM trees and 0.51 (SD = .43) for LMMs. The effects of the most important predictors of predictive accuracy are depicted in Fig. 10. As Fig. 10 indicates, LMM trees show highest predictive accuracy in datasets with purely piecewise interactions, whereas LMMs show highest predictive accuracy in datasets with purely continuous interactions. LMM trees perform poorly only when interactions are purely linear, whereas LMMs perform poorly when interactions are not purely continuous. Strikingly, Fig. 10 suggests that LMMs perform somewhat more accurately in the presence of purely piecewise interactions than in the presence of partly continuous interactions, but only with larger sample sizes and a smaller number of potential moderator variables.
Performance of both LMM trees and LMMS improves with increasing sample size. Furthermore, performance of LMM trees is not affected by the number of covariates, whereas the predictive accuracy of LMMs deteriorates when the number of covariates increases, especially when the true interactions are not purely continuous. This indicates that LMM trees are especially useful for exploratory purposes, where there are many potential moderator variables. In addition, LMM trees may often provide simpler models: Whereas the LMMs included 12.30 significant terms on average, LMM trees had 3.38 inner nodes on average, requiring only about 3–4 variables to be evaluated for making predictions.
Application: Individual patient-level meta-analysis on treatments for depression
Method
Dataset
To illustrate the use of GLMM trees in real data applications, we employ a dataset from an individual-patient data meta-analysis of Cuijpers et al. (2014). This meta-analysis was based on patient-level observations from 14 RCTs, comparing the effects of psychotherapy (cognitive behavioral therapy; CBT) and pharmacotherapy (PHA) in the treatment of depression. The study of Cuijpers et al. (2014) was aimed at establishing whether gender is a predictor or moderator of the outcomes of psychological and pharmacological treatments for depression. Treatment outcomes were assessed by means of the 17-item Hamilton Rating Scale for Depression (HAM-D; Hamilton, 1960). Cuijpers et al. (2014) found no indication that gender predicted or moderated treatment outcome.
In our analyses, post-treatment HAM-D score was the outcome variable, and potential partitioning variables were age, gender, level of education, presence of a comorbid anxiety disorder at baseline, and pre-treatment HAM-D score. The predictor variable in the linear model was treatment type (0 = CBT and 1 = PHA). An indicator for study was used as the cluster indicator.
In RCTs, ANCOVAs are often employed, to linearly control post-treatment values on the outcome measure for pre-treatment values. Therefore, post-treatment HAM-D scores, controlled for the linear effects of pre-treatment HAM-D scores were taken as the outcome variable. All models were fitted using data of the 694 patients from seven studies, for which complete data was available. Results of our analysis may therefore not be fully representative of the complete dataset of the meta-analysis by Cuijpers et al. (2014).
Models and comparisons
As the outcome variable is continuous, we employed an identity link and Gaussian response distribution. The resulting GLMM trees will therefore be referred to as LMM trees. To compare the accuracy of LMM trees, we also fitted LM trees and LMMs with pre-specified interactions to the data. In the LMMs, the outcome variable was regressed on a random intercept, main effects of treatment and the potential moderators (partitioning variables) and interactions between treatment and the potential moderators. As it is not known in advance how to interact the potential moderators, higher-order interactions were not included.
Effect size
To provide a standardized estimate of the treatment effect differences in the final nodes of the trees, we calculated node-specific Cohen’s d values. Cohen’s d was calculated by dividing the node-specific predicted treatment outcome difference by the node-specific pooled standard deviation.
Predictive accuracy
Predictive accuracy of each method was assessed by calculating the average correlation between observed and predicted HAM-D post-treatment scores, based on 50-fold cross validation.
Stability
The results of recursive partitioning techniques are known to be potentially unstable, in the sense that small changes in the dataset may substantially alter the variables or values selected for partitioning. Therefore, following Philipp, Zeileis, & Strobl (2016), subsampling is used to assess the stability of the selected splitting variables and values. More precisely, variable selection frequencies of the trees are computed from 500 subsamples, each comprising 90% of the full dataset.
Results
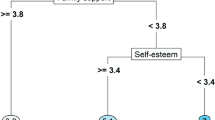
Figures 11 and 12 present the LM and LMM trees fitted on the IPDMA data. The LM tree (Fig. 11) selected level of education as the first partitioning variable, and presence of a comorbid anxiety disorder as a second partitioning variable. By taking into account study-specific intercepts, the LMM tree (Fig. 12) indicates that the first split in the LM tree may be spurious and selected presence of a comorbid anxiety disorder as the only partitioning variable. The terminal nodes of Fig. 12 show a single treatment-subgroup interaction: for patients without a comorbid anxiety disorder, CBT and PHA provide more or less the same reduction in HAM-D scores (Cohen’s d = 0.05). For patients with a comorbid anxiety disorder, PHA provides a greater reduction in HAM-D scores (Cohen’s d = 0.39). The estimated intraclass correlation coefficient for the LMM tree was .05.
LM tree for prediction of treatment outcomes in the IPDMA dataset. The selected partitioning variables are level of education and the presence of a comorbid anxiety disorder. Upper terminal nodes: y-axes represent post-treatment HAM-D scores, x-axes represent treatment levels (cognitive behavior therapy, CBT vs. pharmacotherapy, PHA). Lower terminal nodes represent subgroup-specific descriptive statistics
LMM tree for prediction of treatment outcomes in the IPDMA dataset. The selected partitioning variable is the presence of a comorbid anxiety disorder. Upper terminal nodes: y-axes represent post-treatment HAM-D scores, x-axes represent treatment levels (cognitive behavior therapy, CBT vs. pharmacotherapy, PHA). Lower terminal nodes represent subgroup-specific descriptive statistics
The study-specific distributions of educational level and treatment outcome may explain why the LMM tree did not select level of education as a partitioning variable. Most (55) of the 74 observations with level of education ≤ 1 were part of a single study, which showed a markedly lower mean level of education (M = 2.57, S D = 1.02; 128 observations) compared to the other studies (M = 3.78, S D = 0.53; 566 observations), as well as a markedly higher mean level of post-treatment HAM-D scores (M = 11.20, S D = 6.87) compared to the other studies (M = 7.78, S D = 5.95).
The LMM with pre-specified treatment interactions yielded three significant predictors of treatment outcome: like in the LMM tree, an effect of the presence of a comorbid anxiety disorder was found (main effect: b = 2.29, p = 0.002; interaction with treatment: b = −2.10, p = 0.028). Also, the LMM indicated an interaction between treatment and age (b = .10, p = 0.018).
Assessment of predictive accuracy by means of 50-fold cross validation indicated better predictive accuracy for the LMM tree than for the LM tree and the LMM. The correlation between true and predicted post-treatment HAM-D total scores averaged over 50 folds was .272 (S D = .260) for LMM tree, .233 (S D = .252) for the LMM with pre-specified interactions and .190 (S D = .290) for the LM tree.
Table 3 presents statistics on the variables selected for partitioning in subsamples of the dataset. Presence of a comorbid anxiety disorder was selected for partitioning in the majority of LMM trees grown on subsamples of the dataset, while the other variables were selected in at most 4% of the subsamples. As the comorbid anxiety disorder variable involved only a single splitting value, further assessment of the stability of splitting values was not necessary.
Discussion
Summary
We presented the GLMM tree algorithm, which allows for estimation of a GLM-based recursive partition, as well as estimation of global random-effects parameters. We hypothesized GLMM trees to be well suited for the detection of treatment-subgroup interactions in clustered datasets. We confirmed this through our simulation studies and by applying the algorithm to an existing dataset on the effects of depression treatments.
Our simulations focused on the performance of the GLMM tree algorithm in datasets with continuous response variables. The resulting LMM trees accurately recovered the subgroups in 90% of simulated datasets with treatment-subgroup interactions, outperforming LM trees without random effects. In terms of predictive accuracy, LMM trees outperformed LM trees as well as MERTs, predicting treatment-effect differences in test data with .94 accuracy, on average. In datasets without treatment-subgroup effects, LMM trees showed a rather low type I error rate of 4%, compared with a type I error rate of 33% for LM trees.
Better performance of LMM trees was mostly observed when random effects were sizeable and correlated with potential partitioning variables. In those circumstances, LM trees are likely to detect spurious splits and subgroups. Especially with smaller effect and sample sizes (i.e., Cohen’s d = .5 and/or N = 200), LMM trees outperformed the other tree methods. As such effect and sample sizes are quite common in multi-center clinical trials, the GLMM tree algorithm may provide a useful tool for subgroup detection in those instances.
In the absence of random effects, LM and LMM trees yielded very similar predictive accuracy. This finding is of practical importance, indicating that application of the GLMM tree algorithm will not reduce accuracy when in fact, random effects are absent from the data.
Compared to LMMs with pre-specified interactions, LMM trees provided somewhat better accuracy, on average. LMM trees performed much better than LMMs when interactions were at least partly piecewise. However, the performance of LMM trees deteriorated when the interactions were purely continuous functions of the predictor variables, in which case LMMs with pre-specified interactions performed very well. The performance of LMMs deteriorated with a larger number of predictor variables, whereas the performance of LMM trees was not affected by this, confirming our expectation that LMM trees are better suited for exploration than LMMs.
In the Application, we found the LMM tree yielded somewhat higher predictive accuracy, while using a smaller number of variables than the LM tree and the LMM with pre-specified interactions. The LMM trees obtained over repeated subsamples of the training data proved to be relatively stable.
Limitations and future directions
Recursive partitioning methods were originally developed as a non-parametric tool for classification and regression, assuming the mechanism that generated the data unknown (e.g., Breiman, 2001). The GLMM tree algorithm takes MOB trees and GLMMs as building blocks and thereby inherits some of their sensitivity to model misspecifications.
As with GLMMs, misspecification of the random effects may negatively affect the accuracy of GLMM trees. Previous research on GLMMs has shown that misspecifying the shape of the random-effects distribution reduces the accuracy of random-effects predictions, but has little effects on the fixed-effects parameter estimates (e.g., Neuhaus, McCulloch, & Boylan, 2013). This indicates that for GLMM trees, misspecification of the shape of the random-effects distribution will affect random-effects predictions, but will have little effect on the estimated tree. The incorrect specification of a predictor as having a fixed instead of a random effect may be more likely to result in spurious splits or poorer parameter estimates. Therefore, to check for model misspecification, users are advised to visually inspect residuals and predicted random effects. The tutorial included in the supplementary material shows how residuals and random effects predictions can be obtained and plotted.
As with MOB trees, GLMM trees will perform best when the true data structure is tree-shaped. Trees may need many splits to approximate structures with a different shape, which may be more unstable under small perturbations. Therefore, in the Application we have shown how tree stability can be assessed using the methods developed by Philipp et al. (2016). Furthermore, when relevant partitioning variables are omitted, GLMM tree can only approximate the subgroups using the specified variables. Although our simulations did not involve scenarios where relevant partitioning variables were omitted, the results of Frick, Strobl, and Zeileis (2014) indicate that MOB trees can detect parameter instability even when covariates are only loosely connected to the group structure.
GLMM trees rely more on user specification of predictor variables than methods like CART and MERT. On the one hand, this increases the danger of model misspecification. On the other hand, it increases power and accuracy if the fixed-effects predictor variable(s) are correctly specified. For example, the best approach for assessing treatment-effect differences with MERTs is not completely obvious. We tried two approaches in our simulations: firstly, fitting a single MERT with the treatment indicator as one of the potential partitioning variables and secondly, fitting separate MERTs for each level of the treatment indicator. With the first approach, MERTs often did not detect the treatment indicator as a predictor variable in datasets with treatment-subgroup interactions. The second approach yielded better accuracy, indicating that MERTs profit from user specification of the relevant predictor variable(s). The GLMM tree algorithm provides a straightforward approach for including relevant fixed-effects predictor variables in the model. Our results suggest this yields higher predictive accuracy if the model is correctly specified. However, our simulations have not assessed the effect of misspecification of the fixed-effects predictor variable(s) and such misspecification may reduce predictive accuracy.
For recursive partitioning methods fitting local parametric models only, the danger of model misspecification may be limited (e.g., Ciampi, 1991; Siciliano, Aria, & D’Ambrosio, 2008; Su, Wang, & Fan, 2004). GLMM trees, however, fit a global random-effects model in addition to local fixed-effects regression models and may therefore be less robust against model misspecification. For example, the global random-effects model assumes a single random-effects variance across terminal nodes. If the random-effects variance, however, does vary across terminal nodes, this may negatively affect the performance of the parameter stability tests. Further research on the effects of model misspecifications on the performance of GLMM trees is therefore needed.
In the Introduction, we mentioned several existing tree-based methods for treatment-subgroup interaction detection. These methods have different objectives and there is not yet an agreed-upon single best method. In a simulation study, Sies and Van Mechelen (2016) found the method of Zhang et al. (2012a) to perform best, followed by MOB. However, the method of Zhang et al. performed worst under some conditions of the simulation study in terms of the type I error rate. Further research comparing tree-based methods for treatment-subgroup interaction detection is needed, especially for clustered datasets, as our simulations and comparisons mostly focused on LMM trees and LM trees.
Conclusions
Our results indicate that GLMM trees provide accurate recovery of treatment-subgroup interactions and prediction of treatment effects, both in the presence and absence of random effects and interactions. Therefore, GLMM trees offer a promising method for detecting treatment-subgroup interactions in clustered datasets, for example in multi-center trials or individual-level patient data meta-analyses.
References
Athey, S., & Imbens, G. (2016). Recursive partitioning for heterogeneous causal effects. Proceedings of the National Academy of Sciences, 113(27), 7353–7360.
Bates, D., Mächler, M., Bolker, B., & Walker, S. (2015). Fitting linear mixed-effects models using lme4. Journal of Statistical Software, 67(1), 1–48. https://doi.org/10.18637/jss.v067.i01.
Bates, D., Maechler, M., Bolker, B., Walker, S., Christensen, R., Singmann, H., & Green, P. (2017). Linear Mixed-Effects Models using ’Eigen’ and S4 [Computer software manual]. Retrieved from https://CRAN.R-project.org/package=lme4 (R package version 1.1-7).
Breiman, L. (2001). Statistical modeling: The two cultures (with comments and a rejoinder by the author). Statistical Science, 16(3), 199–231.
Breiman, L., Friedman, J., Olshen, R., & Stone, C. (1984). Classification and Regression Trees. New York: Wadsworth.
Bryk, A. S., & Raudenbush, S. W. (1992). Hierarchical linear models: Applications and data analysis methods. Newbury Park: Sage.
Ciampi, A. (1991). Generalized regression trees. Computational Statistics & Data Analysis, 12(1), 57–78.
Cohen, J. (1992). A power primer. Psychological Bulletin, 112(1), 155–159.
Cooper, H., & Patall, E. A. (2009). The relative benefits of meta-analysis conducted with individual participant data versus aggregated data. Psychological Methods, 14(2), 165.
Cuijpers, P., Weitz, E., Twisk, J., Kuehner, C., Cristea, I., David, D., & Hollon, S. D. (2014). Gender as predictor and moderator of outcome in cognitive behavior therapy and pharmacotherapy for adult depression: An individual-patients data meta-analysis. Depression and Anxiety, 31(11), 941–951.
Doove, L. L., Dusseldorp, E., Van Deun, K., & Van Mechelen, I. (2014). A comparison of five recursive partitioning methods to find person subgroups involved in meaningful treatment-subgroup interactions. Advances in Data Analysis and Classification, 8, 403–425.
Driessen, E., Smits, N., Dekker, J., Peen, J., Don, F. J., Kool, S., & Van, H. L. (2016). Differential efficacy of cognitive behavioral therapy and psychodynamic therapy for major depression: A study of prescriptive factors. Psychological Medicine, 46(4), 731–744.
Dusseldorp, E., & Meulman, J. J. (2004). The regression trunk approach to discover treatment covariate interaction. Psychometrika, 69(3), 355–374.
Dusseldorp, E., & Van Mechelen, I. (2014). Qualitative interaction trees: A tool to identify qualitative treatment-subgroup interactions. Statistics in Medicine, 33(2), 219–237.
Dusseldorp, E., Doove, L., & Van Mechelen, I. (2016). Quint: An R package for the identification of subgroups of clients who differ in which treatment alternative is best for them. Behavior Research Methods, 48, 650.
Fokkema, M., & Zeileis, A. (2016). glmertree: Generalized linear mixed model trees. Retrieved from http://R-Forge.R-project.org/R/?group_id=261 (R package version 0.1-1).
Foster, J. C., Taylor, J. M. G., & Ruberg, S. J. (2011). Subgroup identification from randomized clinical trial data. Statistics in Medicine, 30(24), 2867–2880.
Frick, H., Strobl, C., & Zeileis, A. (2014). To split or to mix? tree vs. mixture models for detecting subgroups. In COMPSTAT 2014-Proceedings in Computational Statistics (pp. 379–386).
Hajjem, A., Bellavance, F., & Larocque, D. (2011). Mixed effects regression trees for clustered data. Statistics & Probability Letters, 81(4), 451–459.
Hamilton, M. (1960). A rating scale for depression. Journal of Neurology, Neurosurgery and Psychiatry, 23(1), 56.
Higgins, J., Whitehead, A., Turner, R. M., Omar, R. Z., & Thompson, S. G. (2001). Meta-analysis of continuous outcome data from individual patients. Statistics in Medicine, 20(15), 2219–2241.
Hothorn, T., & Zeileis, A. (2015). partykit: A modular toolkit for recursive partytioning in R. Journal of Machine Learning Research, 16, 3905–3909. Retrieved from http://www.jmlr.org/papers/v16/hothorn15a.html.
Hothorn, T., & Zeileis, A. (2016). A Toolkit for Recursive Partytioning [Computer software manual]. Retrieved from https://CRAN.R-project.org/package=partykit (R package version 1.1-0).
Kraemer, H. C., Frank, E., & Kupfer, D. J. (2006). Moderators of treatment outcomes: Clinical, research, and policy importance. Journal of the American Medical Association, 296(10), 1286–1289.
Kuznetsova, A., Brockhoff, P., & Christensen, R. (2016). lmertest: Tests in linear mixed effects models [Computer software manual]. Retrieved from https://CRAN.R-project.org/package=lmerTest (R package version 2.0-32).
Lipkovich, I., Dmitrienko, A., Denne, J., & Enas, G. (2011). Subgroup identification based on differential effect search - A recursive partitioning method for establishing response to treatment in patient subpopulations. Statistics in Medicine, 30(21), 2601–2621.
Martin, D. (2015). Efficiently exploring multilevel data with recursive partitioning (Unpublished doctoral dissertation). University of Virginia.
Neuhaus, J. M., McCulloch, C. E., & Boylan, R. (2013). Estimation of covariate effects in generalized linear mixed models with a misspecified distribution of random intercepts and slopes. Statistics in Medicine, 32(14), 2419–2429.
Philipp, M., Zeileis, A., & Strobl, C. (2016). A toolkit for stability assessment of tree-based learners. In Colubi, A., Blanco, A., & Gatu, C. (Eds.) Proceedings of COMPSTAT 2016 – 22nd international conference on computational statistics (pp. 315–325). Oviedo: The International Statistical Institute/International Association for Statistical Computing.
R Core Team (2016). R: A language and environment for statistical computing [Computer software manual]. Vienna, Austria. Retrieved from https://www.R-project.org/.
Seibold, H., Zeileis, A., & Hothorn, T. (2016). Model-based recursive partitioning for subgroup analyses. International Journal of Biostatistics, 12(1), 45–63. https://doi.org/10.1515/ijb-2015-0032.
Sela, R. J., & Simonoff, J. S. (2011). Reemtree: Regression trees with random effects [Computer software manual]. (R package version 0.90.3).
Sela, R. J., & Simonoff, J. S. (2012). RE-EM trees: A data mining approach for longitudinal and clustered data. Machine Learning, 86(2), 169–207.
Siciliano, R., Aria, M., & D’Ambrosio, A. (2008). Posterior prediction modelling of optimal trees. In Compstat 2008 (pp. 323–334). Heidelberg: Physica-Verlag HD.
Sies, A., & Van Mechelen, I. (2016). Comparing four methods for estimating tree-based treatment regimes. (Submitted).
Strobl, C., Malley, J., & Tutz, G. (2009). An introduction to recursive partitioning: Rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychological Methods, 14(4), 323.
Su, X., Wang, M., & Fan, J. (2004). Maximum likelihood regression trees. Journal of Computational and Graphical Statistics, 13(3), 586–598.
Su, X., Tsai, C.-L., Wang, H., Nickerson, D. M., & Li, B. (2009). Subgroup analysis via recursive partitioning. The Journal of Machine Learning Research, 10, 141–158.
Zeileis, A., & Hornik, K. (2007). Generalized M-fluctuation tests for parameter instability. Statistica Neerlandica, 61(4), 488–508.
Zeileis, A., Hothorn, T., & Hornik, K. (2008). Model-based recursive partitioning. Journal of Computational and Graphical Statistics, 17(2), 492–514.
Zhang, B., Tsiatis, A. A., Davidian, M., Zhang, M., & Laber, E. (2012a). Estimating optimal treatment regimes from a classification perspective. Stat, 1(1), 103–114.
Zhang, B., Tsiatis, A. A., Laber, E. B., & Davidian, M. (2012b). A robust method for estimating optimal treatment regimes. Biometrics, 68(4), 1010–1018.
Acknowledgements
The authors would like to thank Prof. Pim Cuijpers, Prof. Jeanne Miranda, Dr. Boadie Dunlop, Prof. Rob DeRubeis, Prof. Zindel Segal, Dr. Sona Dimidjian, Prof. Steve Hollon and Dr. Erica Weitz for granting access to the dataset for the application. The work for this paper was partially done while MF, AZ and TH were visiting the Institute for Mathematical Sciences, National University of Singapore in 2014. The visit was supported by the Institute.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix
Appendix
Glossary
Abbreviations
- CART::
-
Classification and regression trees.
- GLM::
-
Generalized linear model.
- GLMM::
-
Generalized linear mixed model.
- IPDMA::
-
Individual patient data meta analysis.
- LM::
-
Linear model.
- LMM::
-
Linear mixed model.
- MERT::
-
Mixed-effects regression tree.
- ML::
-
Maximum likelihood.
- MOB::
-
Model-based recursive partitioning.
- RECPAM::
-
Recursive partition and amalgamation.
- REML::
-
Restricted maximum likelihood.
Notation
- β j :
-
column vector of fixed-effects coefficients in terminal node j
- b m :
-
column vector of random-effects coefficients in cluster m
- d j :
-
standardized mean difference of treatment outcome in terminal node j
- 𝜖 :
-
deviation of observed treatment outcome y from its expected value
- 1,…,i,…,N :
-
index for observation
- 1,…,j,…,J :
-
index for terminal node
- 1,…,k,…,K :
-
index for partitioning variable
- 1,…,m,…,M :
-
index for cluster
- r :
-
index for iteration
- U k :
-
(potential) partitioning variable k
- x i :
-
column vector of fixed-effects predictor variable values for observation i
- y i :
-
treatment outcome for observation i
- z i :
-
column vector of random-effects predictor variable values for observation i
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Fokkema, M., Smits, N., Zeileis, A. et al. Detecting treatment-subgroup interactions in clustered data with generalized linear mixed-effects model trees. Behav Res 50, 2016–2034 (2018). https://doi.org/10.3758/s13428-017-0971-x
Published:
Issue Date:
DOI: https://doi.org/10.3758/s13428-017-0971-x