Abstract
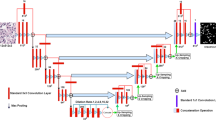
Segmenting nuclei from histopathology images is a crucial step in the early identification and diagnosis of several diseases. Due to the complexity of histopathology images, accurate nucleus segmentation is not a simple operation. However, convolutional neural networks (CNNs) have recently been revealed to be a viable option. The well-known CNN model, namely the U-Net, demonstrated its image segmentation effectiveness in medical field. However, U-Net has several drawbacks, such as information loss after transmission through particular steps. Another significant one is the likelihood of feature mismatches in the encoder and decoder sub-networks in skip connection, which can lead to the fusing of semantically unrelated information and, as a consequence, fuzzy feature maps throughout the learning process. In order to solve these issues, an improved U-Net architecture called Information Added U-Net with Sharp Block (IASB-U-Net) has been proposed for nuclei segmentation from histopathology images. Information is added to the encoder-decoder path in the proposed model after each layer, and sharpening spatial filters are utilized in place of skip connections. The experimental study over a merged dataset demonstrates that the proposed IASB-U-Net produces competitive results when compared to established CNN models such as U-Net, Dense U-Net, SCPP Net, and LiverNet.






Similar content being viewed by others
DATA AVAILABILITY STATEMENT
The authors do not have the permission to share the data.
REFERENCES
Madabhushi, A., Digital pathology image analysis: opportunities and challenges, Imaging Med., 2019, vol. 1, no. 1, p. 7.
Madabhushi, A. and George, L., Image Analysis and Machine Learning in Digital Pathology: Challenges and Opportunities, 2016, pp. 170–175.
Ronneberger, O., Fischer, P., and Brox, T., U-net: Convolutional networks for biomedical image segmentation, in International Conference on Medical image computing and computer-assisted intervention, Springer, Cham, 2015, pp. 234–241.
Zunair, H. and Hamza, A.B., Sharp U-Net: Depthwise convolutional network for biomedical image segmentation, Comput. Biol. Med., 2021, vol. 136, p. 104699.
Lagree, A., Mohebpour, M., Meti, N., Saednia, K., Lu, F.I., Slodkowska, E., Gandhi, S., Rakovitch, E., Shenfield, A., Sadeghi-Naini, A., and Tran, W.T., A review and comparison of breast tumor cell nuclei segmentation performances using deep convolutional neural networks, Sci. Rep., 2021, vol. 11, no. 1, pp.1–11.
Kumar, N., Verma, R., Sharma, S., Bhargava, S., Vahadane, A., and Sethi, A., A dataset and a technique for generalized nuclear segmentation for computational pathology, IEEE Trans. Med, Imaging, 2017, vol. 36, no. 7, pp. 1550–1560.
Kumar, N., Verma, R., Anand, D., Zhou, Y., Onder, O.F., Tsougenis, E., Chen, H., Heng, P.A., Li, J., Hu, Z., and Wang, Y., A multi-organ nucleus segmentation challenge, IEEE Trans. Med. Imaging, 2019, vol. 39, no. 5, pp. 1380–1391.
Naylor, P., Laé, M., Reyal, F., and Walter, T., Segmentation of nuclei in histopathology images by deep regression of the distance map, IEEE Trans. Med. Imaging, 2018, vol. 38, no. 2, pp. 448–459.
Ali, M.A., Misko, O., Salumaa, S.O., Papkov, M., Palo, K., Fishman, D., and Parts, L., Evaluating very deep convolutional neural networks for nucleus segmentation from brightfield cell microscopy images, SLAS Discovery, 2021, vol. 26, no. 9, pp. 1125–1137.
Fishman, D., Salumaa, S.O., Majoral, D., Peel, S., Wildenhain, J., Schreiner, A., Palo, K., and Parts, L., Segmenting nuclei in brightfield images with neural networks, 2019, bioRxiv, p. 764894.
Shuvo, M.B., Ahommed, R., Reza, S., and Hashem, M.M.A., CNL-UNet: A novel lightweight deep learning architecture for multimodal biomedical image segmentation with false output suppression, Biomed. Signal Process. Control, 2021, vol. 70, p. 102959.
Jaeger, S., Karargyris, A., Candemir, S., Folio, L., Siegelman, J., Callaghan, F., Xue, Z., Palaniappan, K., Singh, R.K., Antani, S., and Thoma, G., Automatic tuberculosis screening using chest radiographs, IEEE Trans. Med. Imaging, 2013, vol. 33, no. 2, pp. 233–245.
Candemir, S., Jaeger, S., Palaniappan, K., Musco, J.P., Singh, R.K., Xue, Z., Karargyris, A., Antani, S., Thoma, G., and McDonald, C.J., Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration, IEEE Trans. Med. Imaging, 2013, vol. 33, no. 2, pp. 577–590.
Codella, N., Rotemberg, V., Tschandl, P., Celebi, M.E., Dusza, S., Gutman, D., Helba, B., Kalloo, A., Liopyris, K., Marchetti, M., and Kittler, H., Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic), arXiv preprint arXiv:1902.03368, 2019.
Tschandl, P., Rosendahl, C., and Kittler, H., The HAM10000 dataset, a large collection of multi-sourcedermatoscopic images of common pigmented skin lesions, Sci. Data, 2018, vol. 5, no. 1, pp. 1–9.
Caicedo, J.C., Goodman, A., Karhohs, K.W., Cimini, B.A., Ackerman, J., Haghighi, M., Heng, C., Becker, T., Doan, M., McQuin, C., and Rohban, M., Nucleus segmentation across imaging experiments: the 2018 Data Science Bowl, Nat. Methods, 2019, vol. 16, no. 12, pp. 1247–1253.
2018 data science bowl, https://www.kaggle.com/c/data-science-bowl-2018, 2018.
Ultrasound nerve segmentation, https://www.kaggle.com/c/ultrasound-nerve-segmentation, 2016.
Buda, M., Saha, A., and Mazurowski, M.A., Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm, Comput. Biol. Med., 2019, vol. 109, pp. 218–225.
Buda, M., Brain mri segmentation. 2020, January 10. [Online]. Available: https://www.kaggle.com/mateuszbuda/lgg-mri-segmentation.
Dinh, T.L., Kwon, S.G., Lee, S.H., and Kwon, K.R., Breast tumor cell nuclei segmentation in histopathology images using EfficientUnet++ and Multi-organ transfer learning, J. Korea Multimedia Soc., 2021, vol. 24, no. 8, pp. 1000–1011.
Kadia, D.D., Alom, M.Z., Burada, R., Nguyen, T.V., and Asari, V.K., R2U3D: Recurrent Residual 3D U-Net for Lung Segmentation, 2021. arXiv preprint arXiv:2105.02290.
VESSEL12–Home. Accessed November 4, 2020. [Online]. Available: https://vessel12.grand-challenge.org/.
LUNA16–Home. Accessed November 4, 2020. [Online]. Available: https://luna16.grand-challenge.org/.
Chanchal, A.K., Lal, S., and Kini, J., High-resolution deep transferred ASPPU-Net for nuclei segmentation of histopathology images, Int. J. Comput. Assisted Radiol. Surg., 2021, pp. 1–17.
Irshad, H., Montaser-Kouhsari, L., Waltz, G., Bucur, O., Nowak, J.A., Dong, F., Knoblauch, N.W., and Beck, A.H., Crowdsourcing image annotation for nucleus detection and segmentation in computational pathology: evaluating experts, automated methods, and the crowd, in Pacific symposium on biocomputing Co-chairs, 2014, pp. 294–305.
Yu, H., Fan, D., and Song, W., GPU-Net: Lightweight U-Net with more diverse features, 2022. arXiv preprint arXiv:2201.02656.
Yan, X., Tang, H., Sun, S., Ma, H., Kong, D., and Xie, X., After-unet: Axial fusion transformer unet for medical image segmentation, in Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, 2022, pp. 3971–3981.
Xuming Chen, Shanlin Sun, Narisu Bai, Kun Han, Qianqian Liu, Shengyu Yao, Hao Tang, Chupeng Zhang, Zhipeng Lu, Qian Huang, Guoqi Zhao, Yi Xu, Tingfeng Chen, XiaohuiXie, and Yong Liu, A deep learning-based autosegmentation system for organs-at-risk on whole-body computed tomography images for radiation therapy, Radiother. Oncol., 2021, vol. 160, pp. 175–184.
Landman, B., Zhoubing Xu, Juan Eugenio Igelsias, Styner, M., Langerak, T.R., and Klein, A., 2015 Miccai Multi-Atlas Labeling Beyond the Cranial Vault—Workshop and Challenge.
Lambert, Z., Petitjean, C., Dubray, B., and Ruan, S., Segthor: Segmentation of thoracic organs at risk in ct images, 2019.
Guo, C., Szemenyei, M., Yi, Y., Wang, W., Chen, B., and Fan, C., Sa-unet: Spatial attention u-net for retinal vessel segmentation, In 2020 25th International Conference on Pattern Recognition (ICPR), IEEE, 2021, pp. 1236–1242.
Ali, Y., Janabi-Sharifi, F., and Beheshti, S., Echocardiographic image segmentation using deep Res-U network, Biomed. Signal Process. Control, 2021, vol. 64, pp. 102248.
Ahamed, M.A., Hossain, M.A., and Al Mamun, M., Semantic segmentation of self-supervised dataset and medical images using combination of u-net and neural ordinary differential equations, in 2020 IEEE Region 10 Symposium (TENSYMP), IEEE, 2020, pp. 238–241.
Janowczyk, A. and Madabhushi, A., Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases, J. Pathol. Inf., 2016, p. 7.
Cai, S., Tian, Y., Lui, H., Zeng, H., Wu, Y., and Chen, G., Dense-UNet: A novel multiphoton in vivo cellular image segmentation model based on a convolutional neural network, Quant. Imaging Med. Surg., 2020, vol. 10, no. 6, p. 1275.
Chanchal, A.K., Kumar, A., Lal, S., and Kini, J., Efficient and robust deep learning architecture for segmentation of kidney and breast histopathology images, Comput. Electr. Eng., 2021, vol. 92, p. 107177.
Aatresh, A.A., Alabhya, K., Lal, S., Kini, J., and Saxena, P.P., LiverNet: efficient and robust deep learning model for automatic diagnosis of sub-types of liver hepatocellular carcinoma cancer from H&E stained liver histopathology images, Int. J. Comput. Assisted Radiol. Surg., 2021, pp. 1–15.
Diederik P. Kingma and Jimmy Ba, Adam: A method for stochastic optimization, Yoshua Bengio and Yann Le Cun, Ed., in 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, May 7-9, 2015, Conference Track Proceedings, 2015.
Gudhe, N.R., Behravan, H., Sudah, M., Okuma, H., Vanninen, R., Kosma, V.M., and Mannermaa, A., Multi-level dilated residual network for biomedical image segmentation, Sci. Rep., 2021, vol. 11, no. 1, pp. 1–18.
Kanadath, A., Jothi, J.A.A., and Urolagin, S., Histopathology Image Segmentation Using MobileNetV2 based U-net Model, in 2021 International Conference on Intelligent Technologies (CONIT), IEEE, 2021, pp. 1–8.
Basu, A., Senapati, P., Deb, M., Rai, R., and Dhal, K.G., A survey on recent trends in deep learning for nucleus segmentation from histopathology images, Evol. Syst., 2023, pp. 1–46. https://doi.org/10.1007/s12530-023-09491-3
Deb, M., Garai, A., Das, A., and Dhal, K.G., LS-Net: A convolutional neural network for leaf segmentation of rosette plants, Neural Comput. Appl., 2022, vol. 34, no. 21, pp. 18511–18524.
Deb, M., Dhal, K.G., Mondal, R., and Gálvez, J., Paddy disease classification study: A deep convolutional neural network approach, Opt. Mem. Neural Networks, 2021, vol. 30, pp. 338–357.
Funding
This work was supported by ongoing institutional funding. No additional grants to carry out or direct this particular research were obtained.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This work does not contain any studies involving human and animal subjects.
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest.
Additional information
Publisher’s Note.
Allerton Press remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Anusua Basu, Deb, M., Das, A. et al. Information Added U-Net with Sharp Block for Nucleus Segmentation of Histopathology Images. Opt. Mem. Neural Networks 32, 318–330 (2023). https://doi.org/10.3103/S1060992X23040070
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S1060992X23040070