Abstract
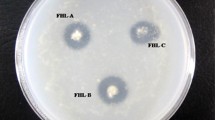
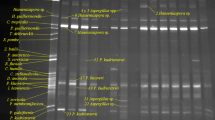
Protease enzymes (proteases), particularly those produced by microorganisms, play very important roles in industry, due to their diverse applications. Considering the richness of microbial diversity in nature, a good chance always exists that proteases more suitable, with better properties for commercial application, may be discovered while screening novel microorganisms from local environments. In this study, 94 yeasts were isolated from different natural sources collected from the Abha region, Kingdom of Saudi Arabia, to determine extracellular protease production and activity. Among them, 23 isolates (24.46%) showed protease activity using a casein hydrolysis test. Of these, five isolates (21.74%) were selected and identified as the best protease producers by exhibiting the largest clearance zones around colonies. A 26S rRNA gene D1/D2 domain sequence alignment, comparison, and phylogenetic analysis of our study yeasts to published D1/D2 domain rRNA gene sequences from GenBank, identifies the isolates as Rhodotorula mucilaginosa KKU-M12c, Cryptococcus albidus KKU-M13c, Pichia membranifaciens KKU-M18c, Hanseniaspora uvarum KKU-M19c, and Candida californica KKU-M20c. The influence of varying pH (4.0–9.0) on the yield and activity of the proteases was investigated using 0.5% (w/v) casein as a substrate, to detect optimum pH values for yeast extracellular protease production. Enzyme activity was measured using qualitative and quantitative assays. Results show all of the study yeasts secreting protease enzyme at all tested pH levels, with the exception of pH 9.0. This indicates that none of the five yeasts are alkaline protease producers. Maximum protease activity (187 U/mL) was observed in strain H. uvarum KKU-M19c at pH 6.0 (only), indicating that strain KKU-M19c only produces neutral protease. The other four yeast isolates, R. mucilaginosa KKU-M12c, C. albidus KKU-M13c, P. membranifaciens KKU-M18c, and C. californica KKU-M20c, produced both acidic (at pH 4.0) and neutral (at pH 6.0 and 7.0) proteases. Strain C. californica KKU-M20c was found to be the best acidic and neutral protease producer (138 U/mL at pH 4.0, and 185 U/mL at pH 7.0). This is the first report of the discovery and isolation of local, powerful yeasts producing acidic and neutral protease enzymes from the Abha region, Kingdom of Saudi Arabia.
Similar content being viewed by others
References
Abirami, V., Meenakshi, S.A., Kanthymathy, K., Bharathidasan, R., Mahalingam, R., and Panneerselvam, A, Partial purification and characterization of an extracellular protease from Penicillium janthinellum and Neurospora crassa, Eur. J. Exp. Biol., 2011, vol. 1, no. 3, pp. 114–123.
Sevinc, N. and Demirkan, E., Production of protease by Bacillus sp. N-40 isolated from soil and its enzymatic properties, J. Biol. Environ. Sci., 2011, vol. 5, no. 14, pp. 95–103.
Gupta, A., Roy, I., Patel, R.K., Singh, S.P., Khare, S.K., and Gupta, M.N., One-step purification and characterization of an alkaline protease from haloalkaliphilic Bacillus sp., J. Chromatogr. A, 2005, vol. 1075, nos. 1–2, pp. 103–108.
El-Shafei, H., Abdel-Aziz, M., Ghaly, M., and Abdalla, A.A.H, Optimizing some factors affecting alkaline protease production by a marine bacterium Streptomyces albidoflavus, in Proc. 5th Sci. Environ. Conf. Egypt. Zagazig Univ., 2010, pp. 125–142.
Haddar, A., Hmidet, N., Bellaaj, O.G., Fakhfakh-Zouari, N., Sellami-Kamoun, A., and Nasri, M, Alkaline proteases produced by Bacillus licheniformis RP1 grown on shrimp wastes: application in chitin extraction, chicken feather degradation and as a dehairing agent, Biotechnol. Bioprocess Eng., 2011, vol. 16, no. 4, pp. 669–678.
Haile, G. and Gessesse, A, Properties of alkaline protease C45 produced by alkaliphilic Bacillus sp. isolated from Chitu, Ethiopian Soda Lake, J. Biotechnol. Biomater., 2012, vol. 2, p. 136.
Wu, T.Y., Mohammad, A.W., Jahim, J.M., and Anuar, N, Investigations on protease production by a wild-type Aspergillus terreus strain using diluted retentate of pre-filtered palm oil mill effluent (POME) as substrate, Enzyme Microb. Technol., 2006, vol. 39, no. 6, pp. 1223–1229.
Krishna, K., Gupta, M., Gupta, N., Gaudani, H., Trivedi, S., Patil, P., Gupta, G., Khairnar, Y., Borasate, A., and Mishra, D, Optimization of growth and production of protease by Penicillium species using submerged fermentation, Int. J. Microbiol. Res., 2009, vol. 1, no. 1, pp. 14–18.
Braga, A.A., de Morais, P.B., and Linardi, V.R, Screening of yeast from brazilian amazon rain forest for extracellular proteinases production, Syst. Appl. Microbiol., 1998, vol. 21, no. 3, pp. 353–359.
Chi, Z., Ma, C., Wang, P., and Li, H.F, Optimization of medium and cultivation conditions for alkaline protease production by the marine yeast Aureobasidium pullulans, Biores. Technol., 2007, vol. 98, no. 3, pp. 534–538.
Kim, J.Y, Isolation of protease-producing yeast Pichia farinosa CO-2 and characterization of its extracellular enzyme, J. Korean Soc. Appl. Biol. Chem., 2010, vol. 53, no. 2, pp. 133–141.
Habib, S.M.A., Fakhruddin, A.N.M., Begum, S., and Ahmed, M.M, Isolation and screening of thermo stable extracellular alkaline protease producing bacteria from tannery effluents, J. Sci. Res., 2012, vol. 4, no. 2, p. 515.
Ogrydziak, D.M, Production of alkaline extracellular protease by Yarrowia lipolytica, Crit. Rev. Biotechnol., 1988, vol. 8, no. 3, pp. 177–187.
Ramakrishna, T. and Pandit, M.W., Self-association of a-chymotrypsin: effect of amino acids, J. Biosci., 1988, vol. 13, no. 3, pp. 215–222.
Hesham, A., New safety and rapid method for extraction of genomic DNA from bacteria and yeast strains suitable for PCR amplifications, J. Pure Appl. Microbiol., 2014, vol. 8, no. 1, pp. 383–388.
Kurtzman, C.P. and Robnett, C.J, Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences, Antonie van Leeuwenhoek, 1998, vol. 73, no. 4, pp. 331–371.
Rao, M.B., Tanksale, A.M., Ghatge, M.S., and Deshpande, V.V, Molecular and biotechnological aspects of microbial proteases, Microbiol. Mol. Biol. Rev., 1998, vol. 62, no. 3, pp. 597–635.
Rowlands, R.T, Industrial strain improvement: mutagenesis and random screening procedures, Enzyme. Microbial. Technol., 1984, vol. 6, no. 1, pp. 3–10.
Gupta, R., Beg, Q.K., and Lorenz, P, Bacterial alkaline proteases: molecular approaches and industrial applications, Appl. Microbiol. Biotechnol., 2002, vol. 59, no. 1, pp. 15–32.
Alnahdi, H.S, Isolation and screening of extracellular proteases produced by new isolated Bacillus sp, J. App. Pharm. Sci., 2012, vol. 2, no. 9, pp. 071–074.
Kurtzman, C.P. and Robnett, C.J, Molecular relationships among hyphal ascomycetous yeasts and yeast-like taxa, Can. J. Bot., 1995, vol. 73, no. 1, pp. 824–830.
Hesham, A., Khan, S., Liu, X., Zhang, Y., Wang, Z., and Yang, M, Application of PCR-DGGE to analyse the yeast population dynamics in slurry reactors during degradation of polycyclic aromatic hydrocarbons in weathered oil, Yeast, 2006, vol. 23, no. 12, pp. 879–887.
Hong, S.G., Chun, J., Oh, H.W., and Bae, K.S, Metschnikowia koreensis sp. nov. A novel yeast species isolated from flowers in Korea, Int. J. Syst. Evol. Microbiol., 2001, vol. 51, pp. 1927–1931.
Herzberg, M., Fischer, R., and Titze, A, Conflicting results obtained by RAPD-PCR and large subunit rDNA sequences in determining and comparing yeast strains isolated from flowers: a comparison of two methods, Int. J. Syst. Evol. Microbiol., 2002, vol. 52, pp. 1423–1433.
Daniel, H.M. and Meyer, W, Evaluation of ribosomal RNA and actin gene sequences for the identification of ascomycetous yeasts, Int. J. Food Microbiol., 2003, vol. 86, nos. 1–2, pp. 61–78.
Frutos, R.L., Fernandez-Espinar, M.T., and Querol., A, Identification of species of the genus Candida by analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers, Antonie van Leeuwenhoek, 2004, vol. 85, no. 3, pp. 175–185.
Kurtzman, C.P, Four new yeasts in the Pichia anomala clade, Int. J. Syst. Evol. Microbiol., 2000, vol. 50, pp. 395–404.
Abliz, P., Fukushima, K., Takizawa, K., and Nishimura, K, Identification of pathogenic dematiaceous fungi and related taxa based on large subunit ribosomal DNA D1/D2 domain sequence analysis, FEMS Immunol. Med. Microbiol., 2004, vol. 40, no. 1, pp. 41–49.
Rodarte, M., Dias, D., Vilela, D., and Schhwan, R, Proteolytic activities of bacteria, yeasts and filamentous fungi isolated from coffee fruit (Coffea arabica L.), Acta Sci. Agron., 2011, vol. 33, no. 3, pp. 457–464.
Kamada, M., Oda, K., and Murao, S, Extracellular protease of yeasts. 3. Purification of the extracellular acid protease of Rhodotorula glutinis K-24 and its general properties, Agric. Biol. Chem., 1972, vol. 36, no. 7, pp. 1095–1101.
Maddox, I.S. and Hough, J.S, Proteolytic enzymes of Saccharomyces carlsbergensis, Biochem. J., 1970, vol. 117, no. 5, pp. 843–852.
Koelsch, G., Tang, J., Loy, J.A., Monod, M., Jackson, K., Foundling, S.I., and Lin, X, Enzymic characteristics of secreted aspartic proteases of Candida albicans, Biochim. Biophys. Acta, 2000, vol. 1480, nos. 1–2, pp. 117–131.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
About this article
Cite this article
Hesham, A.E.L., Alrumman, S.A., Al-Dayel, M.A. et al. Screening and genetic identification of acidic and neutral protease-producing yeasts strains by 26S rRNA gene sequencing. Cytol. Genet. 51, 221–229 (2017). https://doi.org/10.3103/S0095452717030033
Received:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0095452717030033