Abstract
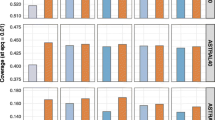
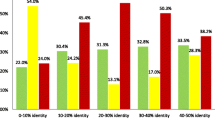
Pairwise alignment of amino acid sequences is the basic tool of bioinformatics, which is widely used both independently and within numerous more complex methods. The effectiveness of this tool critically depends on the scoring function used, which consists of a substitution matrix and gap penalties. In this work, amino acid substitution matrices for the superfamily of microbial rhodopsins (RHOD) were constructed and analyzed and then compared with a set of general-purpose matrices (BLOSUM, VTML, PFASUM). It was shown that all matrices allow constructing alignments of microbial rhodopsin sequences of almost the same quality, but only BLOSUM and VTML matrices and their linear combinations with RHOD matrices allow revealing homology between microbial rhodopsins and heliorhodopsin.
Similar content being viewed by others
References
Govorunova, E.G., Sineshchekov, O.A., Li, H., and Spudich, J.L., Microbial rhodopsins: Diversity, mechanisms, and optogenetic applications, Annu. Rev. Bio-chem., 2017, vol. 86,no. 1, pp. 845–872.
Pushkarev, A., Inoue, K., Larom, S., et al., A distinct abundant group of microbial rhodopsins discovered using functional metagenomics, Nature, 2018, vol. 558,no. 7711, pp. 595–599.
Needleman, S.B. and Wunsch, C.D., A general method applicable to the search for similarities in the amino acid sequence of two proteins, J. Mol. Biol., 1970, vol. 48,no. 3, pp. 443–453.
Smith, T.F. and Waterman, M.S., Identification of common molecular subsequences, J. Mol. Biol., 1981, vol. 147,no. 1, pp. 195–197.
Löytynoja, A., Alignment methods: Strategies, challenges, benchmarking, and comparative overview, in Evolutionary Genomics. Methods in Molecular Biology (Methods and Protocols), Anisimova, M., Ed., New Jersey: Humana Press, 2012, vol. 855, pp. 203–235.
Khan, F.I., Wei, D.Q., Gu, K.R., Hassan, M.I., and Tabrez, S., Current updates on computer aided protein modeling and designing, Int. J. Biol. Macromol., 2016, vol. 85, pp. 48–62.
Thompson, J.D., Higgins, D.G., and Gibson, T.J., Clustal, W: Improving the sensitivity of progressive multiple sequence aligment through sequence weighting, position specific gap penalties and weight matrix choice, Nucleic Acid Res., 1994, vol. 22,no. 22, pp. 4673–4680.
Kuznetsov, I.B., Protein sequence alignment with family-specific amino acid similarity matrices, BMC Res. Notes, 2011, vol. 4, p. 296.
Henikoff, S. and Henikoff, J.G., Amino acid substitution matrices from protein blocks, Proc. Natl. Acad. Sci. U.S.A., 1992, vol. 89,no. 22, pp. 10915–10919.
Müller, T., Spang, R., and Vingron, M., Estimating amino acid substitution models: A comparison of Day-hoff’s estimator, the resolvent approach and a maximum likelihood method, Mol. Biol. Evol., 2002, vol. 19,no. 1, pp. 8–13.
The UniProt Consortium, UniProt: The universal protein knowledgebase, Nucleic Acids Res., 2018, vol. 46,no. 5, pp. 2699–2699.
Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E., The protein data bank, Nucleic Acid Res., 2000, vol. 28,no. 1, pp. 235–242.
Keul, F., Hess, M., Goesele, M., and Hamacher, K., PFASUM: A substitution matrix from Pfam structural alignments, BMC Bioinf., 2017, vol. 18, p. 293.
Pearson, W.R., Rapid and sensitive sequence comparison with FASTP and FASTA, Methods Enzymol., 1990, vol. 183, pp. 63–98.
Sievers, F. and Higgins, D.G., Clustal Omega for making accurate alignments of many protein sequences, Protein Sci., 2018, vol. 27,no. 1, pp. 135–145.
Edgar, R.C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput, Nucleic Acid Res., 2004, vol. 32,no. 5, pp. 1792–1797.
Stamm, M., Staritzbichler, R., Khafizov, K., and Forrest, L.R., AlignMe—a membrane protein sequence alignment web server, Nucleic Acid Res., 2014, vol. 42,no. W1, pp. W246–W251.
Kolbe, M., Besir, H., Essen, L.O., and Oesterhelt, D., Structure of the light-driven chloride pump halorho-dopsin at 1.8 Å resolution, Science, 2000, vol. 288,no. 5470, pp. 1390–1396.
Kouyama, T., Kanada, S., Takeguchi, Y., Narusawa, A., Murakami, M., and Ihara, K., Crystal structure of the light-driven chloride pump halorhodopsin from Natronomonas pharaonis, J. Mol. Biol., 2010, vol. 396,no. 3, pp. 564–579.
Gushchin, I., Reshetnyak, A., Borshchevskiy, V., Ishchenko, A., Round, E., Grudinin, S., Engelhard, M., Büldt, G., and Gordeliy, V., Active state of sensory rho-dopsin II: Structural determinants for signal transfer and proton pumping, J. Mol. Biol., 2011, vol. 412,no. 4, pp. 591–600.
Kato, H.E., Inoue, K., Abe-Yoshizumi, R., et al., Structural basis for Na+ transport mechanism by a light-driven Na+ pump, Nature, 2015, vol. 521,no. 7550, pp. 48–53.
Finn, R.D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R.Y., Eddy, S.R., Heger, A., Hethe-rington, K., Holm, L., Mistry, J., Sonnhammer, E.L.L., Tate, J., and Punta, M., Pfam: The protein families database, Nucleic Acid Res., 2014, vol. 42,no. D1, pp. D222–D230.
Zhang, J., Mizuno, K., Murata, Y., Koide, H., Murakami, M., Ihara, K., and Kouyama, T., Crystal structure of deltarhodopsin-3 from Haloterrigena ther-motolerans, Proteins: Struct., Funct., Bioinf., 2013, vol. 81,no. 9, pp. 1585–1592.
Sadovnichy, V., Tikhonravov, A., Voevodin, V., and Opanasenko, V.I., “Lomonosov:” Supercomputing at Moscow State University, in Contemporary High Performance Computing: From Petascale toward Exascale, Boca Raton: CRC Press, 2013, pp. 283–307.
Author information
Authors and Affiliations
Corresponding author
Additional information
Russian Text © The Author(s), 2019, published in Vestnik Moskovskogo Universiteta, Seriya 16: Biologiya, 2019, Vol. 74, No. 1, pp. 27–33.
About this article
Cite this article
Novoseletsky, V.N., Armeev, G.A. & Shaitan, K.V. Construction and Analysis of Amino Acid Substitution Matrices for Optimal Alignment of Microbial Rhodopsin Sequences. Moscow Univ. Biol.Sci. Bull. 74, 21–25 (2019). https://doi.org/10.3103/S009639251901005X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S009639251901005X