Abstract
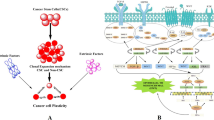
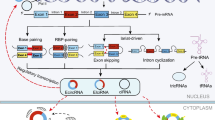
Current therapies against metastatic tumors are still ineffective. Cancer stem cells — a small subset of cells inside the tumor that possesses a self-renewal capacity — might be responsible for the recurrence of the tumor after anti-cancer therapies. Their immortality and unique drug resistance impede their eradication during therapy. The ‘stemness’ of these cells is controlled by microRNAs. These molecules possess the ability to downregulate gene expression by binding to the target mRNA. It turns out that microRNAs control the expression of approximately 60% of the genes in human cells. MicroRNA aberrant expression can lead to cancer development and progression. Therefore, recent research has focused on unraveling the role of microRNA in maintaining a stem-like phenotype in malignant tumors and cancer stem cells. This review summarizes our current knowledge about microRNAs that control the self-renewal capacity of cancer stem cells and indicates the importance of profound research aimed at developing efficient miRNA-targeted therapies.
Similar content being viewed by others
References
Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., et al., MicroRNA genes are transcribed by RNA polymerase II, EMBO J., 2004, 23, 4051–4060
Cai X., Hagedorn C.H., Cullen B.R., Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs, RNA, 2004, 10, 1957–1966
Borchert G.M., Lanier W., Davidson B.L., RNA polymerase III transcribes human microRNAs, Nat. Struct. Mol. Biol., 2006, 13, 1097–10101
Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., et al., The Microprocessor complex mediates the genesis of microRNAs, Nature, 2004, 432, 235–240
Yi R., Qin Y., Macara I.G., Cullen B.R., Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs, Genes Dev., 2003, 17, 3011–3016
Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J., Processing of primary microRNAs by the Microprocessor complex, Nature, 2004, 432, 231–235
Jagadeeswaran G., Zheng Y., Sumathipala N., Jiang H., Arrese E.L., Soulages J.L., et al., Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development, BMC Genomics, 2010, 11, 52
Zhang L., Ding L., Cheung T.H., Dong M.Q., Chen J., Sewell A.K., et al., Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2, Mol. Cell, 2007, 28, 598–613
Babiarz J.E., Ruby J.G., Wang Y., Bartel D.P., Blelloch R., Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs, Genes Dev., 2008, 22, 2773–2785
Berezikov E., Liu N., Flynt A.S., Hodges E., Rooks M., Hannon G.J., et al., Evolutionary flux of canonical microRNAs and mirtrons in Drosophila, Nat. Genet., 2010, 42, 6–9
Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A., Identification of mammalian microRNA host genes and transcription units, Genome Res., 2004, 14, 1902–1910
Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E., mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes, Genes Dev., 2006, 20, 1885–1898
Bartel D.P., MicroRNAs: target recognition and regulatory functions, Cell, 2009, 136, 215–233
Eulalio A., Huntzinger E., Izaurralde E., Getting to the root of miRNA-mediated gene silencing, Cell, 2008, 132, 9–14
Chekulaeva M., Filipowicz W., Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells, Curr. Opin. Cell Biol., 2009, 21, 452–460
Ørom U.A., Nielsen F.C., Lund A.H., MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation, Mol. Cell, 2008, 30, 460–471
Henke J.I., Goergen D., Zheng J., Song Y., Schüttler C.G., Fehr C., et al., microRNA-122 stimulates translation of hepatitis C virus RNA, EMBO J., 2008, 27, 3300–3310
Reya T., Morrison S.J., Clarke M.F., Weissman I.L., Stem cells, cancer, and cancer stem cells, Nature, 2001, 414, 105–111
Clarke M.F., Dick J.E., Dirks P.B., Eaves C.J., Jamieson C.H., Jones D.L., et al., Cancer stem cells—perspectives on current status and future directions: AACR Workshop on cancer stem cells, Cancer Res., 2006, 66, 9339–9344
Gupta P.B., Chaffer C.L., Weinberg R.A., Cancer stem cells: mirage or reality? Nat. Med., 2009, 15, 1010–1012
Welte Y., Adjaye J., Lehrach H.R., Regenbrecht C.R., Cancer stem cells in solid tumors: elusive or illusive? Cell Commun. Signal., 2010, 8, 6
Liu S., Dontu G., Wicha M.S., Mammary stem cells, self-renewal pathways, and carcinogenesis, Breast Cancer Res., 2005, 7, 86–95
Dontu G., Jackson K.W., McNicholas E., Kawamura M.J., Abdallah W.M, Wicha M.S., Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells, Breast Cancer Res, 2004, 6, R605–R615
Karhadkar S.S., Bova G.S., Abdallah N., Dhara S., Gardner D., Maitra A., et al., Hedgehog signalling in prostate regeneration, neoplasia and metastasis, Nature, 2004, 431, 707–712
Olsen C.L., Hsu P.P., Glienke J., Rubanyi G.M., Brooks A.R., Hedgehog-interacting protein is highly expressed in endothelial cells but down-regulated during angiogenesis and in several human tumors, BMC Cancer, 2004, 4, 43
Nam Y., Aster J.C., Blacklow S.C., Notch signaling as a therapeutic target, Curr. Opin. Chem. Biol., 2002, 6, 501–509
Nickoloff B.J., Osborne B.A., Miele L., Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents, Oncogene, 2003, 22, 6598–6608
Bjerkvig R., Tysnes B.B., Aboody K.S., Najbauer J., Terzis A.J., Opinion: the origin of the cancer stem cell: current controversies and new insights, Nat. Rev. Cancer, 2005, 5, 899–904
Nowell P.C., The clonal evolution of tumor cell populations, Science, 1976, 194, 23–28
Ailles L.E., Weissman I.L., Cancer stem cells in solid tumors, Curr. Opin. Biotechnol., 2007, 18, 460–466
Bonnet D., Dick J.E., Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell, Nat. Med., 1997, 3, 730–737
Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., et al., A cell initiating human acute myeloid leukaemia after transplantation into SCID mice, Nature, 1994, 367, 645–648
Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F., Prospective identification of tumorigenic breast cancer cells, Proc. Natl. Acad. Sci. U.S.A., 2003, 100, 3983–3988
Ponti D., Costa A., Zaffaroni N., Pratesi G., Petrangolini G., Coradini D., et al., Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties, Cancer Res., 2005, 65, 5506–5511
Dontu G., Abdallah W.M., Foley J.M., Jackson K.W., Clarke M.F., Kawamura M.J., et al., In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells, Genes Dev., 2003, 17, 1253–1270
O’Brien C.A., Pollett A., Gallinger S., Dick J.E., A human colon cancer cell capable of initiating tumour growth in immunodeficient mice, Nature, 2007, 445, 106–110
Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., et al., Identification of human brain tumour initiating cells, Nature, 2004, 432, 396–401
Suetsugu A., Nagaki M., Aoki H., Motohashi T., Kunisada T., Moriwaki H., Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells, Biochem. Biophys. Res. Commun., 2006, 351, 820–824
Fang D., Nguyen T.K., Leishear K., Finko R., Kulp A.N., Hotz S., et al., A tumorigenic subpopulation with stem cell properties in melanomas, Cancer Res., 2005, 65, 9328–9337
Richardson G.D., Robson C.N., Lang S.H., Neal D.E., Maitland N.J., Collins A.T., CD133, a novel marker for human prostatic epithelial stem cells, J. Cell Sci., 2004, 117, 3539–3545
Maeda S., Shinchi H., Kurahara H., Mataki Y., Maemura K., Sato M., et al., CD133 expression is correlated with lymph node metastasis and vascular endothelial growth factor-C expression in pancreatic cancer, Br. J. Cancer, 2008, 98, 1389–1397
Monzani E., Facchetti F., Galmozzi E., Corsini E., Benetti A., Cavazzin C., et al., Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential, Eur. J. Cancer, 2007, 43, 935–946
Jin L., Hope K.J., Zhai Q., Smadja-Joffe F., Dick J.E., Targeting of CD44 eradicates human acute myeloid leukemic stem cells, Nat. Med., 2006, 12, 1167–1174
Collins A.T., Berry P.A., Hyde C., Stower M.J., Maitland N.J., Prospective identification of tumorigenic prostate cancer stem cells, Cancer Res., 2005, 65, 10946–10951
Li C., Heidt D.G., Dalerba P., Burant C.F., Zhang L., Adsay V., et al., Identification of pancreatic cancer stem cells, Cancer Res., 2007, 67, 1030–1037
Quintana E., Shackleton M., Foster H.R., Fullen D.R., Sabel M.S., Johnson T.M., et al., Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized, Cancer Cell., 2010, 18, 510–523
Sztiller-Sikorska M., Koprowska K., Jakubowska J., Zalesna I., Stasiak M., Duechler M., et al., 2012. Sphere formation and self-renewal capacity of melanoma cells is affected by the microenvironment, Melanoma Res., 2010, 22, 215–224
Pinner S., Jordan P., Sharrock K., Bazley L., Collinson L., Marais R., et al., Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination, Cancer Res., 2009, 69, 7969–7977
Cheli Y., Giuliano S., Botton T., Rocchi S, Hofman V., Hofman P., et al., Mitf is the key molecular switch between mouse or human melanoma initiating cells and their differentiated progeny, Oncogene, 2011, 30, 2307–2318
Du J., Widlund H.R., Horstmann M.A., Ramaswamy S., Ross K., Huber W.E., et al., Critical role of CDK2 for melanoma growth linked to its melanocytespecific transcriptional regulation by MITF, Cancer Cell, 2004, 6, 565–576
Carreira S., Goodall J., Denat L., Rodriguez M., Nuciforo P., Hoek K.S., et al., Mitf regulation of Dia1 controls melanoma proliferation and invasiveness, Genes Dev., 2006, 20, 3426–3439
Park I.K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., et al., Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells, Nature, 2003, 423, 302–305
Molofsky A.V., He S., Bydon M., Morrison S.J., Pardal R., Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways, Genes Dev., 2005, 19, 1432–1437
Pietersen A.M., Evers B., Prasad A.A., Tanger E., Cornelissen-Steijger P., Jonkers J., et al., Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium, Curr. Biol., 2008, 18, 1094–1099
Christoffersen N.R., Silahtaroglu A., Orom U.A., Kauppinen S., Lund A.H., miR-200b mediates post-transcriptional repression of ZFHX1B, RNA, 2007, 13, 1172–1178
Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., et al., The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1, Nat. Cell Biol., 2008, 10, 593–601
Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., et al., Reprogramming of human somatic cells to pluripotency with defined factors, Nature, 2008, 451, 141–146
Simon J.A, Kingston R.E., Mechanisms of polycomb gene silencing: knowns and unknowns, Nat. Rev. Mol. Cell Biol., 2009, 10, 697–708
Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., et al., Polycomb complexes repress developmental regulators in murine embryonic stem cells, Nature, 2006, 441, 349–353
Herranz N., Pasini D., Díaz V.M., Francí C., Gutierrez A., Dave N., et al., Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor, Mol. Cell Biol., 2008, 28, 4772–4781
Hussain M., Rao M., Humphries A.E., Hong J.A., Liu F., Yang M., et al., Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells, Cancer Res., 2009, 69, 3570–3578
Glinsky G.V., Berezovska O., Glinskii A.B., Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer, J. Clin. Invest., 2005, 115, 1503–1521
Iliopoulos D., Lindahl-Allen M., Polytarchou C., Hirsch H.A., Tsichlis P.N., Struhl K., Loss of miR-200 inhibition of Suz12 leads to polycombmediated repression required for the formation and maintenance of cancer stem cells, Mol. Cell, 2010, 39, 761–772
Biddle A., Liang X., Gammon L., Fazil B., Harper L.J., Emich H., et al., Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratoryor proliferative, Cancer Res., 2011 71, 5317–5326
Brabletz T., Jung A., Spaderna S., Hlubek F., Kirchner T., Opinion: migrating cancer stem cells — an integrated concept of malignant tumour progression, Nat. Rev. Cancer, 2005, 5, 744–749
Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., et al., The epithelial-mesenchymal transition generates cells with properties of stem cells, Cell, 2008, 133, 704–715
Thiery J.P., Epithelial-mesenchymal transitions in development and pathologies, Curr. Opin. Cell Biol., 2003, 15, 740–746
Wellner U., Schubert J., Burk U.C., Schmalhofer O., Zhu F., Sonntag A., et al., The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs, Nat. Cell Biol., 2009, 11, 1487–1495
Wang X.Q., Ongkeko W.M., Chen L., Yang Z.F., Lu P., Chen K.K., et al., Octamer 4 (Oct4) mediates chemotherapeutic drug resistance in liver cancer cells through a potential Oct4-AKT-ATP-binding cassette G2 pathway, Hepatology, 2010, 52, 528–539
Frank N.Y., Margaryan A., Huang Y., Schatton T., Waaga-Gasser A.M., Gasser M., et al., ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma, Cancer Res., 2005, 65, 4320–4333
Gupta P.B., Onder T.T., Jiang G., Tao K., Kuperwasser C., Weinberg R.A., et al., Identification of selective inhibitors of cancer stem cells by high-throughput screening, Cell, 2009, 138, 645–659
Stefani G., Slack F.J., Small non-coding RNAs in animal development, Nat. Rev. Mol. Cell Biol., 2008, 9, 219–230
Chen J.F., Murchison E.P., Tang R., Callis T.E., Tatsuguchi M., Deng Z., Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure, Proc. Natl. Acad. Sci. U.S.A., 2008, 105, 2111–2116
Davis T.H., Cuellar T.L., Koch S.M., Barker A.J., Harfe B.D., McManus M.T., et al., Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus, J. Neurosci., 2008, 28, 4322–4330
Koralov S.B., Muljo S.A., Galler G.R., Krek A., Chakraborty T., Kanellopoulou C., et al., Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage, Cell, 2008, 132, 860–874
Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R., DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal, Nat. Genet., 2007, 39, 380–385
Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., et al., The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans, Nature, 2000, 403, 901–906
Wulczyn F.G., Smirnova L., Rybak A., Brandt C., Kwidzinski E., Ninnemann O., et al., Post-transcriptional regulation of the let-7 microRNA during neural cell specification, FASEB J., 2007, 21, 415–426
Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., et al., Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival, Cancer Res., 2004, 64, 3753–3756
Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., et al., let-7 regulates self renewal and tumorigenicity of breast cancer cells, Cell, 2007, 131, 1109–1123
Johnson C.D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D., et al., The let-7 microRNA represses cell proliferation pathways in human cells, Cancer Res., 2007, 67, 7713–7722
Shimono Y., Zabala M., Cho R.W., Lobo N., Dalerba P., Qian D., et al., Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells, Cell, 2009, 138, 592–603
Godlewski J., Nowicki M.O., Bronisz A., Williams S., Otsuki A., Nuovo G., et al., Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal, Cancer Res., 2008, 68, 9125–9130
Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., et al., A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells, EMBO Rep., 2008, 9, 582–589
Xu N., Papagiannakopoulos T., Pan G., Thomson J.A., Kosik K.S., MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells, Cell, 2009, 137, 647–658
Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., et al., A mammalian microRNA expression atlas based on small RNA library sequencing, Cell, 2007, 129, 1401–1414
Zaman M.S., Chen Y., Deng G., Shahryari V., Suh S.O., Saini S., et al., The functional significance of microRNA-145 in prostate cancer, Br. J. Cancer, 2010, 103, 256–264
Noguchi S., Mori T., Hoshino Y., Yamada N., Nakagawa T., Sasaki N., et al., Comparative study of anti-oncogenic microRNA-145 in canine and human malignant melanoma, J. Vet. Med. Sci., 2012, 74, 1–8
Barroso-del Jesus A., Lucena-Aguilar G., Menendez P., The miR-302-367 cluster as a potential stemness regulator in ESCs, Cell Cycle, 2009, 8, 394–398
Card D.A., Hebbar P.B., Li L., Trotter K.W., Komatsu Y., Mishina Y., et al., Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells, Mol. Cell Biol., 2008, 28, 6426–6238
Lin S.L., Chang D.C., Chang-Lin S., Lin C.H., Wu D.T., Chen D.T., et al., Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state, RNA, 2008, 14, 2115–2124
Ji Q., Hao X., Zhang M., Tang W., Yang M., Li L., et al., MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells, PLoS One, 2009, 4, e6816
Kato M., Paranjape T., Müller R.U., Nallur S., Gillespie E., Keane K., et al., The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells, Oncogene, 2009, 28, 2419–2424
Kozaki K., Imoto I., Mogi S., Omura K., Inazawa J., Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer, Cancer Res., 2008, 68, 2094–2105
Bommer G.T., Gerin I., Feng Y., Kaczorowski A.J., Kuick R., Love R.E., et al., p53-mediated activation of miRNA34 candidate tumor-suppressor genes, Curr. Biol., 2007, 17, 1298–1307
Hermeking H., The miR-34 family in cancer and apoptosis, Cell Death Differ., 2010, 17, 193–199
Liu C., Kelnar K., Liu B., Chen X., Calhoun-Davis T., Li H., et al., The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44, Nat. Med., 2011, 17, 211–215
Furuta M., Kozaki K.I., Tanaka S., Arii S., Imoto I., Inazawa J., miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma, Carcinogenesis, 2010, 31, 766–776
Saini S., Majid S., Yamamura S., Tabatabai L., Suh S.O., Shahryari V., et al., Regulatory Role of mir-203 in Prostate Cancer Progression and Metastasis, Clin. Cancer Res., 2011, 17, 5287–5298
Garzia L., Andolfo I., Cusanelli E., Marino N., Petrosino G., De Martino D., et al., MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma, PLoS One, 2009, 4, e4998
Iorio M.V., Visone R., Di Leva G., Donati V., Petrocca F., Casalini P., et al., MicroRNA signatures in human ovarian cancer, Cancer Res, 2007, 67, 8699–86707
Murakami Y., Yasuda T., Saigo K., Urashima T., Toyoda H., Okanoue T., et al., Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues, Oncogene, 2006, 25, 2537–2545
Zhang J., Luo N., Luo Y., Peng Z., Zhang T., Li S., microRNA-150 inhibits human CD133-positive liver cancer stem cells through negative regulation of the transcription factor c-Myb, Int. J. Oncol., 2012, 40, 747–756
Chen C.Z., Li L., Lodish H.F., Bartel D.P., MicroRNAs modulate hematopoietic lineage differentiation, Science, 2004, 303, 83–86
Ji J., Yamashita T., Budhu A., Forgues M., Jia H.L., Li C., et al., Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells, Hepatology, 2009, 50, 472–480
Clevers H., Wnt/beta-catenin signaling in development and disease, Cell, 2006, 127, 469–480
Wang Y, Yu Y., Tsuyada A., Ren X., Wu X., Stubblefield K., et al., Transforming growth factor-β regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM, Oncogene, 2011, 30, 1470–1480
Yu Z., Wang C., Wang M., Li Z., Casimiro M.C., Liu M., et al., A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation, J. Cell Biol., 2008, 182, 509–517
Hayashita Y., Osada H., Tatematsu Y., Yamada H., Yanagisawa K., Tomida S, et al., A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation, Cancer Res., 2005, 65, 9628–9632
Esquela-Kerscher A., Trang P., Wiggins J.F., Patrawala L., Cheng A., Ford L., et al., The let-7 microRNA reduces tumor growth in mouse models of lung cancer, Cell Cycle, 2008, 7, 759–764
Trang P., Medina P.P., Wiggins J.F., Ruffino L., Kelnar K., Omotola M., et al., Regression of murine lung tumors by the let-7 microRNA, Oncogene, 2010, 29, 1580–1587
Kota J., Chivukula R.R., O’Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.W., et al., Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model, Cell, 2009, 137, 1005–1017
Kumar M.S., Erkeland S.J., Pester R.E., Chen C.Y., Ebert M.S., Sharp P.A., et al., Suppression of non-small cell lung tumor development by the let-7 microRNA family, Proc. Natl. Acad. Sci. U.S.A., 2008, 105, 3903–3908
Ji Q., Hao X., Meng Y., Zhang M., Desano J., Fan D., et al., Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres, BMC Cancer, 2008, 8, 266
Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., et al., Silencing of microRNAs in vivo with ‘antagomirs’, Nature, 2005, 438, 685–689
Gabriely G., Wurdinger T., Kesari S., Esau C.C., Burchard J., Linsley P.S., et al., MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators, Mol. Cell Biol., 2008, 28, 5369–5380
Anand S., Majeti B.K., Acevedo L.M., Murphy E.A., Mukthavaram R., Scheppke L., et al., MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis, Nat. Med., 2010, 16, 909–914
Zheng G., Ambros V., Li W.H., Inhibiting miRNA in Caenorhabditis elegans using a potent and selective antisense reagent, Silence, 2010, 1, 9
Krützfeldt J., Kuwajima S., Braich R., Rajeev K.G., Pena J., Tuschl T., et al., Specificity, duplex degradation and subcellular localization of antagomirs, Nucleic Acids Res., 2007, 35, 2885–2892
Owczarzy R., You Y., Groth C.L., Tataurov A.V., Stability and mismatch discrimination of locked nucleic acid-DNA duplexes, Biochemistry, 2011, 50, 9352–9367
Oh Y.K., Park T.G., siRNA delivery systems for cancer treatment, Adv. Drug Deliv. Rev., 2009, 61, 850–862
Broderick J.A., Zamore P.D., MicroRNA therapeutics, Gene Ther., 2011, 18, 1104–1110
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Wozniak, M. microRNA in the control of stem-like phenotype of cancer cells. cent.eur.j.biol. 8, 931–942 (2013). https://doi.org/10.2478/s11535-013-0222-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11535-013-0222-9