Abstract
Background
In Japan, postoperative relapse occurs within five years in 9.2 to 16% of patients whose breast cancers have not metastasized to lymph nodes at the time of initial surgery (node-negative, n0). Attempts to find molecular markers able to classify n0 breast cancers in terms of postoperative prognosis have not been successful.
Methods
To identify molecular indicators of prognosis for this type of cancer, we used a cDNA microarray consisting of 25,344 human genes to investigate expression profiles of 12 primary breast can-cers from patients whose tumors recurred within five years after surgery (5Y-R) and 12 from patients who survived disease-free for more than five years (5Y-F).
Results
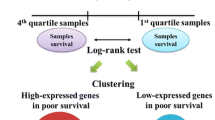
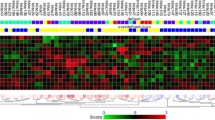
Sets of genes characterizing each group in terms of expression patterns in the tumors were selected by Mann-Whitney and random-permutation tests: these panels included 21 genes expressed highly in 5Y-R tumors than in 5Y-F tumors, and 37 with higher expression in the 5Y-F group than in the 5Y-R group.
Conclusions
We established a scoring system to prediction of postoperative prognosis which was 100% accurate as to the actual clinical outcomes of the 24 cases and therefore might be useful for predict-ing prognosis of n0 breast cancers in a clinical setting. The prognostic score system clearly separated the two groups without any overlap, and accurately predicted prognosis in 6 additional cases. Moreover, the extensive list of tumor-related genes identified in these experiments provides valuable information about progression of breast cancer and suggests potential target molecules for therapy of n0 breast cancers.
Similar content being viewed by others
References
Utada Y, Emi M, Yoshimoto M,et al: Allelic loss at lp34-36 predicts poor prognosis in node-negative breast cancer.Clin Cancer Res 6:3193–3198, 2000.
Saimura M, Fukutomi T, Tsuda H, Utada Y,et al. Prognosis of a series of 763 consecutive node-negative breast cancer patients without adjuvant therapy: Analysis of clinicopathological prognostic factor.J Surg Oncol 71:101–105, 1999.
Utada Y, Haga S, Kajiwara T,et al: Alleile loss at the 8p22 region as a prognostic factor in large and estrogen receptor negative breast carcinoma.Cancer 88:1410–1416, 2000.
Utada Y, Haga S, Kajiwara T,et al: Mapping of target regions of allelic loss in primary breast cancers to 1-cM intervals on genomic contigs at 6q21 and 6q25.3.Jpn J Cancer Res 91:293–300, 2000.
Perou CM, Jeffrey SS, van de Rijn M,et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers.Proc Natl Acad Sci USA 96:9212–9217, 1999.
Perou CM, Sorlie T, Eisen MB,et al: Molecular portraits of human breast tumours.Nature 406:747–752, 2000.
West M, Blanchette C, Dressman H,et al. Predicting the clinical status of human breast cancer by using gene expression profiles.Proc Natl Acad Sci USA 98:11462–11467, 2001.
Gruvberger S, Ringner M, Chen Y, Panavally S,et al: Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns.Cancer Res 61:5979–5984, 2001.
Martin KJ, Kritzman BM, Price LM,et al: Linking gene expression patterns to therapeutic groups in breast cancer.Cancer Res 60:2232–2238, 2000.
Zajchowski DA, Bartholdi MF, Gong Y,et al: Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells.Cancer Res 61:5168–5178, 2001.
Sorlie T, Perou CM, Tibshirani R,et al: Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications.Proc Natl Acad Sci USA 98:10869–10874, 2001.
Abrams JS: Adjuvant therapy for breast cancer-results from the USA consensus conference.Breast Cancer 8:298–304, 2001.
Yang YH, Dudoit S, Luu P,et al: Normalization for cDNA microarray data: a robust compositive method adressing single and multiple slide systematic variation.Nucleic Acids Res 30:el5, 2002.
Manos EJ, Jones DA: Assessment of Tumor Necrosis Factor Receptor and Fas Signaling Pathways by Transcriptional Profiling.Cancer Res 61:433–438, 2001.
Ono K, Tanaka T, Tsunoda T,et al: Identification by cDNA microarray of genes involved in ovarian carcinogenesis.Cancer Res 60:5007–5011, 2000.
Tetsuya S, Saied A, Meng K,et al: Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray.PNAS 97:9127–9123, 2000.
Harris JR, Henderson IC: Staging and prognostic factors. In: Harris, JR. (eds) Breast Diseases. Lippencott publish, Philadelphia, pp327–346, 1987.
McGuire W, Clark G: Prognostic factors and treatment decision in axillary node-negative breast cancers.N Engl J Med 326:1756–1761, 1992.
Michael FP, Leslie B, Patricia AT,et al. HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas.J Clin Oncol 15:2894–2904, 1997.
Elledge RM, Fuqua SAW, Clark GM,et al. Prognostic significance of p53 gene mutations in node-negative breast cancer.Breast Cancer Res Treat 26:225–235, 1993.
Barry I, Fabienne G, Brenda P,et al. Analysis of p53 gene mutation by polymerase chain reaction-single strand conformation polymorphism provides independent prognostic information in node-negative breast cancer.Clin Cancer Res 4:1597–1602, 1998.
Kute TE, Hansen JG, Shao SM,et al: Low cathepsis D and low plasminogen activator type 1 inhibitor in tumor cytosis defines a group of node negative breast cancer patients with low risk of recurrence.Breast Cancer Res Treatment 47:9–16, 1998.
Neilsen NH, Arnerlov C, Emdin SO, Landberg G: CyclinE overexpression, a negative prognostic factor in breast cancer with strong correlation to oestrogen receptor status.Br J Cancer 74:874–880, 1996.
Couzin J: Taking aim at celera’s shotgun.Science 295:1817, 2002.
McPherson JD, Marra M, Hillier L,et al: A physical map of the human genome.Nature 409:934–941, 2001.
Dressman MA, Baras A, Malinowaski R,et al: Geen expression profiling detects gene amplification and differentiates tumor types in breast cancer.Cancer Res 63:2194–2199, 2003.
Unger MA, Rishi M, Clemmer VB,et al: Characterization of adjuvant breast tumors using oligonucleotide microarrays.Breast Cancer Res 3:336–341, 1999.
Assersohn L, Gangi L, Zhao Y,et al: The feasibility of using fine needle aspiration from primary breast cancers for cDNA microarray analysis.Clin Cancer Res 8:794–801, 2002.
van’t Veer LJ, Dai H, van de Vijver MJ,et al: Gene expression profiling predicts clinical outcome of breast cancer.Nature 415:530–536, 2002.
Ahr A, Karn T, Solbach C,et al: Identification of high risk breast-cancer predicts by gene expression profiling.Lancet 359:131–132, 2002.
Lawrence CW, Hinkle DC: DNA polymerase zeta and the control of DNA damage induced mutagenesis in eukaryotes.Cancer Surv 28:21–31, 1996.
Axel H, Hughes L, Nathalie N: Up-regulation of galectin-1 and -3 in human colon cancer and their role in regulating cell and migration.Int J Cancer 103:370–379, 2003.
Zhu F, Jin CX, Song T, Yang J, Cuo L, Yu YN: Response of human REV3 gene to gastric cancer inducing carcinogen N-methyl-N-nitro-N-nitrosoguanidine and its role in mutagenesis.World J Gastroenterol May 9:888–893, 2003.
Ishiduka Y, Mochizuki R, Yanai K,et al: Induction of hydroxyapatite resorptive activity in bone marrow cell populations resistant to bafilomycin Al by a factor with restricted expression to bone and brain, neu-rochondrin.Biochim Biophys Acta 1450:92–98, 1999.
Ishiduka Y, Mochizuki R, Yanai K: Induction of hydroxyapatite resorptive activity in bone marrow cell populations resistant to bafilomycin Al by a factor with restricted expression to bone and brain, neu-rochondrin.Biochim Biophys Acta 1450:92–98, 1999.
Nagahata T, Hirano A, Utada Y,et al: Correlation of alleic losses and clinicopathological factors in 504 primary breast cancers.Breast Cancer 9:208–215, 2002.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Tsumagari, K., Chijiiwa, K., Nagai, H. et al. Postoperative prognosis of node-negative breast cancers predicted by gene-expression profiling on a cDNA microarray of 25,344 genes. Breast Cancer 12, 166–177 (2005). https://doi.org/10.2325/jbcs.12.166
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.2325/jbcs.12.166