Abstract
MicroRNAs (or miRs) play a crucial role in chronic lymphocytic leukemia (CLL) physiopathology and prognosis. In addition, circulating microRNAs in body fluids have been proposed as new biomarkers. We investigated the expression of matched cellular and serum circulating microRNA-150 by quantitative real-time PCR (qPCR) from purified CD19+ cells or from CLL serums obtained at diagnosis in a cohort of 273/252 CLL patients with a median follow-up of 78 months (range 7–380) and correlated it to other biological or clinical parameters. We showed that miR-150 was significantly overexpressed in CLL cells/serums compared with healthy subjects (P < 0.0001). Among CLL patients, a low cellular miR-150 expression level was associated with tumor burden, disease aggressiveness and poor prognostic factors. In contrast, a high level of serum miR-150 was associated with tumor burden markers and some markers of poor prognosis. Similarly, cellular and serum miR-150 also predicted treatment-free survival (TFS) and overall survival (OS) in an opposite manner: patients with low cellular/serum miR-150 levels have median TFS of 40/111 months compared with high-level patients who have a median TFS of 122/60 months (P < 0.0001/P = 0.0066). Similar results were observed for OS. We also found that cellular and serum miR-150 levels vary in an opposite manner during disease progression and that cellular miR-150 could be regulated by its release into the extracellular space. Cellular and serum levels of miR-150 are associated with opposite clinical prognoses and could be used to molecularly monitor disease evolution as a new prognostic factor in CLL.
Similar content being viewed by others
Introduction
Chronic lymphocytic leukemia (CLL), the most common leukemia in Western countries, is characterized by the accumulation of a subtype of CD5+ B lymphocytes in blood, lymph nodes and bone marrow (1). Despite the continuous introduction of new treatments, CLL remains incurable. In addition, CLL displays a very heterogeneous clinical outcome: some patients will rapidly progress and die, whereas others remain asymptomatic for many years (2). These different prognoses can be predicted by several prognostic factors, including clinical markers, such as Binet stage classification; molecular markers, such as mutational status of immunoglobulin heavy-chain variable region (IgHV) and its surrogate markers, ζ-associated protein-70 (ZAP70) and lipoprotein lipase (LPL); serum markers, including β2-microglobulin (β2-M) or soluble CD23 (sCD23); proliferation markers, such as lymphocyte doubling time (LDT); cytogenetic markers; and surface markers, including CD38 molecule (CD38) (reviewed in 3).
In the last decade, several lines of investigation have highlighted the crucial role of microRNAs in the physiopathology of hematological malignancies (4). MicroRNAs (miR) are small, noncoding RNAs of ∼22 nucleotides that can regulate gene expression by hybridizing with a target mRNA sequence, resulting in translational repression or message degradation (5). For all of these reasons, microRNAs have been considered a new class of tumor suppressors (6). More than 200 reports have already shown the importance of microRNAs in the prognosis and the pathogenesis of CLL. Recently, extracellular circulating microRNAs were also detected in body fluids, such as serum, plasma, saliva and urine (7). These circulating microRNAs are believed to be chaperoned by various carriers, such as secreted membrane vesicles (exosomes from 50 to 100 nm and microvesicles from 100 nm to 1 µm) or protein/lipid complexes (8,9), since carrier-free microRNAs will be degraded by RNase digestion and other environmental factors (9). Their levels and composition have been shown to be modulated with injury conditions as well as tumor burden (10–12). For these reasons, extracellular microRNAs could be used as informative biomarkers to assess and monitor disease evolution (11,13).
Moussay et al. (14) recently observed that some circulating microRNAs in CLL plasma could be used as prognostic factors. Among these, microRNA-150 warranted further investigation for a number of reasons. This microRNA plays an important role in normal and malignant hematopoiesis (15). In addition, plasma (14) and cellular (16,17) microRNA-150 were both downregulated in poor prognosis CLL patients based on ZAP70 and IgHV mutational status. A low level of cellular microRNA-150 was also observed in CLL proliferation centers (18), and microRNA-150 is differentially expressed according to the stereotyped B-cell receptor (BCR) subset taken into consideration (19), emphasizing its pivotal role in CLL. Finally, a recent study showed that microRNA-150 was the most abundant microRNA in CLL cells and that it can contribute to the proficiency of BCR signaling (17).
Therefore, in the present report, we extensively studied the prognostic significance of cellular and serum microRNA-150 in a cohort of 273 and 252 CLL patients, respectively. MicroRNA-150 levels were compared with those of normal donors and correlated with several available prognostic factors, clinical data and patient disease evolution.
Materials and Methods
Patients and Sample Collection and Preparation
This study, approved by the Bordet Institute Ethics Committee, was based on 273 peripheral blood samples collected at diagnosis (before any treatment) from CLL patients after written informed consent. All patients had a typical CD19+CD5+ CD23+ phenotype. The median age at diagnosis was 63 years (range 34–90). The median follow-up duration was 78 months (range 7–380). Supplementary Table S1 and Figure S1 summarize other patient characteristics. Control samples were obtained from the peripheral blood of 15 age-matched healthy volunteers (mean 69 years old; range 54–90) after getting written informed consent. Peripheral blood mononuclear cells were isolated by density-gradient centrifugation over Linfosep (Biomedics, Madrid, Spain). B cells were purified with a CD19+ magnetic-bead system (MidiMACS, Miltenyi Biotec, Bergish Gladbash, Germany) according to the manufacturer’s instructions. Mean B-cell purity was >99%, as measured by flow cytometry. Of these 273 patients, 252 serum samples were available. Serum was obtained after a 20-min centrifugation at 300g and was stored at −80°C. All prognostic factors have been measured as previously described (20,21). Details about assessment of prognostic factors can be found in Supplementary Text S1.
RNA Extraction and MicroRNA Quantification
Cellular and serum RNA extraction is described in Supplementary Text S2. For serum RNA, 25 fmol of a mixture of two synthesized Caenorhabditis elegans (cel) microRNA (cel-miR-39 and cel-miR-54; Integrated DNA Technologies, Leuven, Belgium) were spiked in as an exogenous control as previously described (22). MicroRNA expression was measured by using TaqMan microRNA quantitative real-time PCR (qPCR) (Life Technologies [Thermo Fisher Scientific Inc., Waltham, MA, USA]). Briefly, 10 ng total cellular RNA or 5 µL serum RNA was reverse-transcribed by using the MicroRNA Reverse Transcription Kit (Life Technologies [Thermo Fisher Scientific]) and a specific reverse transcription stem-loop primer according to the manufacturer’s protocol. We measured the expression of small nucleolar RNA, C/D box 48 (RNU48) as an endogenous control for cellular miR-150 and microRNA-16, as an endogenous control for serum miR-150 as previously described (23,24). MiR-150/RNU48 and miR-150/miR-16/cel-miR-39/cel-miR-54 were retrotranscribed in the same reaction using 1 µL of each appropriate stem-loop primer (purchased from Life Technologies [Thermo Fisher Scientific]). When cycle threshold (Cq) was >35, the microRNA was considered absent. The expression of each microRNA was normalized with the endogenous or exogenous control and calibrated by subtracting 10 and 5 (chosen arbitrarily) from the ΔCq for serum and cellular miR expression, respectively. The comparative ΔΔCq method was then applied for data analysis, and fold changes were subsequently calculated (fold change = 2−ΔΔCq). All qPCRs were run in duplicate.
Microvesicles and Exosomes Precipitation
To elucidate if circulating microRNA-150 was encapsulated in exosomes, in microvesicles or in protein/lipid complexes, 15 CLL serums were centrifuged at 20,000g (to pellet microvesicles) and subsequently at 150,000g (to pellet exosomes) for 1 h at 4°C as previously described (25,26). Uncentrifuged, microvesicle-depleted and exosome-depleted sera were processed as described above to detect microRNA levels. Because miR-16 and miR-150 could have a different compartmentalization in the serum, miR-150 expression was normalized with the mean of exogenous controls.
Statistical Analysis
We analyzed receiver operating characteristic (ROC) curves with GraphPad Prism 5.0 (Graph-Pad Software) to determine the ZAP70, LPL, CD38, sCD23 and β2-M expression cutoff values that best distinguished IgHV mutated and unmutated cases, considered the gold standard of prognostication. For cellular and serum miR-150 cutoff determination, recursive partitioning to maximize treatment-free survival (TFS)/overall survival (OS) prediction was used. These two techniques for cutoff determination, previously described (23,27), give similar results with some differences because IgHV mutational status does not perfectly predict TFS/OS. The association between microRNA expression and Binet stage was assessed by the Kruskal-Wallis test, and the Mann-Whitney nonparametric test was used for other variables. For paired comparison, the Wilcoxon matched pairs test was used. TFS and OS distributions were plotted by using Kaplan-Meier estimates and were compared using the log-rank test. TFS and OS were calculated from the time of diagnosis until the date of first treatment and the date of death, respectively. All deaths were CLL related. Univariate and multivariate Cox regression analyses were used to evaluate the effects of the different prognostic variables on TFS and OS. All tests were two-sided. An effect was considered to be statistically significant at P < 0.05. All analyses were performed with SPSS 15.0 software.
All supplementary materials are available online at https://doi.org/www.molmed.org.
Results
Cellular and Serum miR-150 Levels Are Upregulated in CLL Compared with Healthy Controls
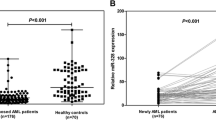
Choice of qPCR normalizers in cells and serum (RNU48 and miR-16) is detailed in Supplemental Text S3 and Supplementary Figures S2–S5. Cellular and serum miR-150 levels in 273 and 252 CLL patients, respectively, were compared with 15 healthy age-matched controls. Cellular miR-150 was expressed 3.6-fold higher in CLL B-cells than in normal purified B cells (P < 0.0001), whereas serum miR-150 level was 7.5-fold higher in CLL sera than in those of healthy volunteers (P < 0.0001) (Figure 1). It should be noted that the levels of miR-150 in the cell and in the serum could not be directly compared because these values are based on different normalizers.
miR-150 is overexpressed in CLL cells and serum compared with healthy controls. Expression of cellular miR-150 (A) from 273 CLL samples (normalized with RNU48) and serum miR-150 (B) from 252 CLL samples (normalized with miR-16) was measured and compared with purified peripheral normal B cells from 15 age-matched healthy volunteers and serum from 15 age-matched healthy volunteers, respectively. Significant differences were assessed by using the Mann-Whitney nonparametric test. The median in the two groups is represented by the red bar.
A Low Level of Cellular miR-150 Is Associated with Higher Tumor Burden, Disease Aggressiveness and Poor Prognostic Factors
The expression of cellular miR-150, as determined by qPCR on purified CD19+ cells and normalized with RNU48, was significantly associated with markers of tumor burden (Binet stage, LDT, sCD23 and β2-M) (Supplementary Table S1 and Figure 2). The level of this microRNA decreased significantly with progression from Binet stage A to C (P = 0.0085). Moreover, patients with an LDT <1 year or who were positive for sCD23 or β2-M expressed significantly lower levels of cellular miR-150. Similar results were obtained for several classic poor prognostic markers commonly used to prognosticate CLL evolution (IgHV mutational status, ZAP70, LPL and CD38 expression, and cytogenetic abnormalities) (Supplementary Table S1 and Figure 2). All of these factors were found to be significant predictors of TFS and OS in our CLL cohort (Supplementary Figure S1).
Low level of cellular miR-150 is associated with higher tumor burden, disease aggressiveness and poor prognostic factors. Cellular miR-15 levels were measured by qPCR and were plotted with their median according to Binet stages (A), untreated/treated patients (B), alive/dead patients (C), β2-M (D), CD23 (E), LDT (F), IgHV mutational status (G), ZAP70 (H), LPL (I), CD38 (J) and cytogenetic abnormalities (K). MicroRNA expression was normalized with RNU48 expression and calibrated by subtracting 5 (chosen arbitrarily) from the ΔCt. The comparative ΔΔCq method was then applied for data analysis, and fold changes were subsequently calculated (fold-change = 2−ΔΔCq). Statistical differences were assessed by using the Kruskal-Wallis test for Binet stages, and the Mann-Whitney nonparametric test was used for other variables. Statistical details can be found in Supplementary Table S1.
A High Level of Serum miR-150 Is Associated with Higher Tumor Burden, with Treatment and/or Death Status and with Some Poor Prognostic Markers
The expression of serum miR-150, as determined by qPCR from serum RNA and normalized with miR-16, was significantly associated with markers of tumor burden (LDT, sCD23 and β2-M) (Supplementary Table S1 and Figure 3). However, in this case, patients with the poorest prognosis (β2-M+, sCD23+, LDT <1 year) had a higher amount of serum miR-150 (P = 0.0017, P < 0.0001, P = 0.0015, respectively). The level of this microRNA was also higher in patients who received a treatment (P < 0.0001), patients who died during the study (P < 0.0001), CD38+ patients (P = 0.0006) and patients with an unfavorable cytogenetic profile (P = 0.0286). Surprisingly, no difference was found according to Binet stage, IgHV, ZAP70 and LPL status (Figure 3).
High level of serum miR-150 is associated with higher tumor burden, treatment/death status and some prognostic markers. Serum miR-15 levels measured by qPCR were plotted with their median according to Binet stages (A), untreated/treated patients (B), alive/dead patients (C), β2-M (D), CD23 (E), LDT (F), IgHV mutational status (G), ZAP70 (H), LPL (I), CD38 (J) and cytogenetic abnormalities (K). MicroRNA expression was normalized with miR-16 expression and calibrated by subtracting 10 (chosen arbitrarily) from the ΔCt. The comparative ΔΔCq method was then applied for data analysis, and fold-changes were subsequently calculated (fold-change = 2−ΔΔCt). Statistical differences were assessed by using the Kruskal-Wallis test for Binet stages, and the Mann-Whitney nonparametric test was used for other variables. Statistical details can be found in Supplementary Table S1.
Cellular and Serum miR-150 Are Opposite Prognostic Factors
The median TFS of the cohort of 273 and 252 patients was 87 months (range 1–267) and 84 months (range 1–267), respectively, whereas the median OS was 237 months (range 1–380) and 242 months (range 1–380), respectively. Using recursive partitioning to maximize TFS/OS prediction, we defined a cutoff for the cellular and serum levels of microRNA-150 (Supplementary Table S1). Among the 273/252 patient cohort, 180 (66%)/133 (53%) were deemed positive and 93 (34%)/119 (47%) were deemed negative for cellular and serum microRNA-150, respectively. Using these cutoffs, cellular and serum miR-150 were predictors of TFS and OS but in an opposite manner: the median TFS of cellular and serum miR-150+ patient subgroups were 111 and 63 months, respectively, whereas these values were 40 and 111 months for the negative subgroups (cellular, P < 0.0001; serum, P = 0.0087). OS was also statistically associated in the log-rank tests with cellular (P = 0.0239) and serum miR-150 (P = 0.0342): the miR-150+ subgroups median OS were >380 and 237 months for cellular and serum values, whereas these values were 159 and >380 months for the negative subgroups, respectively (Figure 4). We also used Cox regression to evaluate the impact of these dichotomized data (by using Supplementary Table S1 cutoffs) on TFS and OS. Cellular and serum miR150 were both univariate predictors of TFS and OS. For cellular miR-150, negative patients had a 2.1-fold higher risk of requiring treatment (P < 0.0001) and a 1.9-fold higher risk of death (P = 0.0262). For serum miR-150, positive patients had a 1.6-fold higher risk of requiring treatment (P = 0.0124) and a 2.1-fold higher risk of death (P = 0.0435) (Supplementary Table S2).
Cellular and serum miR-150 can predict TFS and OS with an opposite manner. (A, D) Representative TFS and OS curves for cellular miR-150 (n = 273). (B, E) Serum miR-150 (n = 252). (C, F) Representative TFS and OS Kaplan-Meier curves, respectively, when status of cellular and serum miR-150 was combined. Cutoffs were optimized by recursive partitioning to maximize TFS/OS (Supplementary Table S1). Statistical differences between curves were calculated using the log-rank test.
Cellular and Serum miR-150 Levels Are Independent from Each Other
Interestingly, we did not find any significant correlation between cellular and serum miR-150 levels (Supplementary Figure S6). In other words, patients deemed positive for cellular miR-150 were not necessarily negative for serum miR-150 and vice versa. Multivariate analysis including cellular and serum miR-150 indicated that both markers confer survival information for TFS and OS prediction (Supplementary Table S2). Therefore, when these two markers were combined, we were able to stratify patients in three distinct subgroups: favorable, intermediate and poor prognosis (Figure 4C).
Multivariate Cox Regression Analysis
In a multivariate stepwise analysis (n = 199) that included seven different factors (IgHV mutational status, ZAP70, LPL, CD38, LDT, β2-M and Binet stage) and cellular and serum miR-150 status, only IgHV mutational status (P < 0.0001), LDT (P < 0.0001) and Binet stage (P = 0.0062) were selected as independent and significant predictors of TFS (Supplementary Table S3). For OS multivariate analysis, ZAP70 (P = 0.0005), CD38 (P = 0.0406) and LDT (P = 0.0064) were selected (Supplementary Table S3). To elucidate if miR-150 is linked to LDT, LDT was removed and Cox regression analysis was run again (n = 227): this time, IgHV (P = 0.0001), serum miR-150 (P = 0.0324) and Binet stage (P < 0.0001) were selected to be significant independent factors, whereas ZAP70 (P = 0.0001) and CD38 (P = 0.0106) were selected for OS (Supplementary Table S4). Although the absolute lymphocyte count (ALC) was statistically associated with cellular and serum miR-150 (Supplementary Figure S7), when the same cox regression analysis (n = 186) was performed including ALC, this last parameter was not selected in the final Cox model. However, serum miR-150 was kept in the model but with a nonsignificant P value (P = 0.0783), indicating that ALC and serum miR-150 are not independent but can provide slightly different prognostic information (Supplementary Table S5). All these data taken together indicated that miR-150 is linked to LDT/ALC for TFS prediction. However, other regression analyses (data not shown) indicated that serum miR-150 is independent from other variables for TFS prediction. Indeed, Supplementary Figure S8 shows that serum miR-150 could identify patients with a worse prognosis in a good prognostic subgroup, such as Binet stage A, IgHV-mutated patients and patients with a better prognosis in the ZAP70-positive subgroup.
Cellular and Serum miR-150 Across Disease Evolution: Relation with Lymphocytosis, Relapse and Remission
Cellular and serum miR-150 levels were investigated in serial samples of the same patients (n = 7). Figure 5A shows disease evolution in relation to lymphocytosis. We observed that overall cellular miR-150 levels decreased when lymphocytosis increased, whereas serum miR-150 was directly proportional to lymphocytosis. It should be noted that even if cellular and serum miR-150 evolution are opposite, no correlation was found when all patients were taken together (R2 =0.0645, P = 0.3610). When cellular or serum miR-150 levels were compared in the same patient at diagnosis and after relapse (Figure 5B), no significant differences were found. Interestingly, after molecular remission, serum miR-150 levels fell to values close to or lower than those of healthy volunteers (Figure 5C).
Cellular and serum miR-150 across disease evolution. (A, B) Evolution of cellular and serum miR-150, respectively, in seven different patients according to lymphocytosis evolution. Cellular (C) and serum (D) miR-150 level were compared in paired samples of the same patient at diagnosis and after relapse (n = 8). (E) Serum miR-150 level was compared in paired samples from the same patients at diagnosis and after molecular remission (n = 7). The Wilcoxon matched pairs nonparametric test was used to calculate significant differences.
The Majority of Serum miR-150 Does Not Circulate in Exosomes/Microvesicles
To further investigate the serum localization of miR-150, specifically whether it is found in microvesicles, in exosomes or free circulating in protein/lipid complexes, sera were depleted of microvesicles (centrifugation at 20,000g) or exosomes (centrifugation at 150,000g). A simple comparison of Cq values indicated that some changes exist after centrifugation (Figures 6A-D). However, very surprisingly, these changes were also observed in the Cq of exogenous controls (cel-miR-39 or cel-miR-54). The same results were obtained in three independent experiments, each including 10–15 samples, indicating that Cq values without normalization and thus efficiency or RNA extraction are affected by the removal of microvesicles or exosomes. Therefore, we decided to normalize data with the Cq of an exogenous control (cel-miR-39 or cel-miR-54 gives similar results). MicroRNA-16 (normalized to the exogenous control) was not used as a normalizer in this experiment because miR-16 itself is influenced by the centrifugation. Indeed, 31.1 ± 4.9% of miR-16 was lost after 20,000g centrifugation, whereas 64.6 ± 3.0% was lost after the removal of exosomes (n = 15, P < 0.0001 and P = 0.0001, respectively) (Figure 6F). For miR-150, 22.2 ± 7.5% of the miR was lost in the microvesicle pellet (P = 0.0054), whereas no statistical loss was observed after the removal of exosomes (28.0 ± 6.0%, P = 0.2126) (Figure 6E). In conclusion, our data taken together indicate that the great majority of miR-150 exists in the serum in an exosome- and microvesicle-free form, most likely associated with protein/lipid complexes.
The majority of serum miR-150 does not circulate in exosomes or microvesicles. A total of 15 CLL serum samples were centrifuged at 20,000g(to pellet microvesicles) and subsequently at 150,000g (to pellet exosomes). Uncentrifuged, vesicle-depleted and exosome-depleted sera were then compared. Cq values of miR-150 (A), miR-16 (B) and exogenous control cel-miR-39 (C) and cel-miR-59 (D) are provided. miR-150 (E) and miR-16 (F) Cq were normalized with the exogenous control cel-miR-39. The mean ratios in the two groups were plotted with their median. The Wilcoxon matched pairs nonparametric test was used to calculate significant differences.
Discussion
The aim of the present study was to evaluate the clinical value of cellular and serum miR-150 because this microRNA is the most abundant microRNA in CLL cells (17) and because its plasma form has been suggested as a biomarker in CLL (14).
Here, we confirmed that miR-150 is overexpressed in CLL cells and serum compared with healthy controls, as was previously reported by Mraz et al. (17) (for its cellular expression) and by Moussay et al. (14) (for its extracellular plasma expression). MicroRNA-150 deregulation has been reported in various cancers, including cutaneous T-cell lymphoma (28) and Burkitt lymphoma (29), as well as in solid cancers, such as in esophagi squamous carcinoma (30), non-small-cell lung carcinoma (31) and colorectal cancer (32). Surprisingly, cellular miR-150 upregulation was not always observed in tumoral tissue when compared with normal tissue; indeed, a lower level of cellular miR-150 is observed in esophagi (30), lung (31) and colorectal cancer (32) and T-cell lymphoma (28). However, when only the tumoral tissue is taken into consideration, low cellular miR-150 expression is always associated with a greater tumoral potential and a poorer prognosis, as confirmed in the present study.
We next showed that cellular miR-150 is significantly decreased in poor prognostic patients (independently of the chosen prognostic factor). These results are in line with those of Fulci et al. (16), who reported a low cellular miR-150 expression in IgHV-unmutated patients, as well as with those of Mraz et al. (17), who observed a low expression in Rai III-IV patients. In addition, this downregulation was reported in the proliferation center of CLL lymph nodes (18). Recently, it was demonstrated that miR-150 could affect BCR signaling and that a lower expression of miR-150 was found in IgM responder patients (17). All of these data taken together indicate that miR-150 may be a potential tumor suppressor and that its downregulation is associated with a better response to microenvironment stimulus that could lead to enhanced cell survival or proliferation and thus a worse patient prognosis.
In this study, we observed for the first time that serum miR-150 is increased in patients with a high tumor burden, CD38+ patients and patients with unfavorable cytogenetic abnormalities. However, we did not find any significant association of serum miR-150 with other prognostic markers, such as ZAP70. This observation disagrees with those of Moussay et al. who reported a lower level of miR-150 in the plasma of poor prognostic patients (according to ZAP70 status). This discrepancy could be explained by several differences between these two studies: the body fluid used (plasma versus serum in our study), number of patients (39 versus 252) and, most importantly, the normalization method (spiked-in exogenous control versus miR-16 endogenous control). Indeed, we showed that microRNA stability can vary with time and that an endogenous control is more suitable for retrospective studies (Supplementary Text S4). Interestingly, we also show that miR-150 serum is linked to lymphocytosis, strengthening the conclusion that a high level in the serum is associated with tumor burden and thus poor prognosis. Another study that evaluated the level of plasma miR-150 in patients undergoing myeloablative chemotherapy followed by autologous hematopoietic stem cell transplantation confirmed this observation (12). Patients requiring treatment also presented a 2.3-fold increase of serum miR-150 compared with untreated patients. Similar results were obtained when we compared surviving and deceased patients, indicating that the high level of serum miR-150 microRNA is linked to disease aggressiveness.
We demonstrated for the first time in a 273/252 patient cohort with a long follow-up that cellular and serum miR-150 could individually predict TFS and OS in an opposite manner: patients with low cellular miR-150 or patients with a high serum miR-150 level had a shorter TFS and OS. However, we could not demonstrate a correlation between cellular and serum miR-150 levels; in other words, patients who were negative for miR-150 cellular expression were not necessarily positive in the serum. This finding could be explained by the fact that other cells could release this microRNA. Because these two parameters were independent, we combined them and were able to separate patients into three prognosis groups. Multivariate analysis including the most commonly used classical factors showed that serum miR-150 linked to LDT but was independent from the other factors for TFS prediction and could also thus refine other prognostic markers such as Binet stage, IgHV mutational status or ZAP70.
Contrary to IgHV mutational status, which does not change during disease evolution, miR-150 is a dynamic factor. Indeed, in serial samples from the same patient, we observed a decrease in cellular miR-150 with disease aggressiveness and increased lymphocytosis, whereas the opposite effect was observed for serum miR-150. However, even after treatment or relapse, we do not see a difference in terms of cellular or serum miR-150 level. Interestingly, after complete molecular remission, serum miR-150 falls back to normal levels. Serum miR-150 could thus be a new tool to molecularly follow disease evolution. However, before use of serum miR-150 level to follow residual disease, other investigations are needed in an independent series of patients to determine the sensitivity of this measure and also an accurate cutoff for residual disease positivity. All these data taken together highly suggested that serum miR-150 is released by circulating CLL cells. Using serums, we were not able to demonstrate a statistical correlation between mir-150 cellular and serum level, probably because other cells could release microRNAs in the extracellular space. By using culture supernatant of purified CLL cells, we were able to show that CLL cells release miR-150 and, in addition, that patients with low levels of cellular miR-150 release higher levels of miR-150 (data not shown). In addition, we showed that this release was not specific to miR-150: indeed, when an irrelevant CLL microRNA was electroporated (as miR-1, which is normally absent in CLL), it was also released in a higher proportion from cellular miR-150 low patients (data not shown). This result is consistent with the fact that low levels of other microRNAs such as miR-34a, mir-29 and miR-223 have been observed in patients with poor prognostic CLL (21,33).
Extracellular microRNA could circulate in body fluids in different forms: encapsulated in microvesicles, in exosomes or associated with protein/lipid complexes. In the present study, by using different speeds of centrifugation, we demonstrated that a small part of miR-150 circulates within microvesicles, but that the majority of circulating miR-150 is most likely associated with protein/lipid complexes because free mi-croRNA will be rapidly degraded by RNAse. These results are in line with the observation of Arroyo et al. (9), who reported that miR-150 is found in both vesicles >120 nm and complexes <1 nm associated with the Ago2 protein.
Conclusion
In conclusion, cellular and serum miR-150 are dynamic prognostic factors in CLL. We have shown that cellular and serum miR-150 levels are associated with the clinical course of CLL in an opposite manner. In addition, serum miR-150 level is an independent predictor of TFS. Our observations indicate that miR-150 determination could help to molecularly monitor disease evolution and will help to better identify high-risk CLL patients who might benefit from earlier treatment.
Disclosure
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
References
Herishanu Y, Katz BZ, Lipsky A, Wiestner A. (2013) Biology of chronic lymphocytic leukemia in different microenvironments: clinical and therapeutic implications. Hematol. Oncol. Clin. North Am. 27:173–206.
Hamblin T. (2002) Chronic lymphocytic leukaemia: one disease or two? Ann. Hematol. 81:299–303.
Van Bockstaele F, Verhasselt B, Philippe J. (2009) Prognostic markers in chronic lymphocytic leukemia: a comprehensive review. Blood Rev. 23:25–47.
Fabbri M, et al. (2008) MicroRNAs and noncoding RNAs in hematological malignancies: molecular, clinical and therapeutic implications. Leukemia. 22:1095–105.
Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–97.
Chen CZ. (2005) MicroRNAs as oncogenes and tumor suppressors. N. Engl. J. Med. 353:1768–71.
Weber JA, et al. (2010) The microRNA spectrum in 12 body fluids. Clin. Chem. 56:1733–41.
Vickers KC, Remaley AT. (2012) Lipid-based carriers of microRNAs and intercellular communication. Curr. Opin. Lipidol. 23:91–7.
Arroyo JD, et al. (2011) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U. S. A. 108:5003–8.
Rosell R, Wei J, Taron M. (2009) Circulating MicroRNA signatures of tumor-derived exosomes for early diagnosis of non-small-cell lung cancer. Clin. Lung Cancer. 10:8–9.
Gilad S, et al. (2008) Serum microRNAs are promising novel biomarkers. PLoS One. 3:e3148.
Pritchard CC, et al. (2012) Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev. Res. (Phila). 5:492–7.
Mitchell PS, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 105:10513–8.
Moussay E, et al. (2011) MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 108:6573–8.
He Y, Jiang X, Chen J. (2014) The role of miR-150 in normal and malignant hematopoiesis. Oncogene. 24;33:3887–93.
Fulci V, et al. (2007) Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 109:4944–51.
Mraz M, et al. (2014) MicroRNA-150 contributes to the proficiency of B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1 genes. Blood. 124:84–95.
Wang M, et al. (2008) miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-150 and high BIC/miR-155 expression. J. Pathol. 215:13–20.
Papakonstantinou N, et al. (2013) Differential microRNA profiles and their functional implications in different immunogenetic subsets of chronic lymphocytic leukemia. Mol. Med. 19:115–23.
Stamatopoulos B, et al. (2007) Quantification of ZAP70 mRNA in B cells by real-time PCR is a powerful prognostic factor in chronic lymphocytic leukemia. Clin. Chem. 53:1757–66.
Stamatopoulos B, et al. (2009) microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 113:5237–45.
McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. (2011) Analysis of circulating microRNA: preanalytical and analytical challenges. Clin. Chem. 57:833–40.
Stamatopoulos B, et al. (2010) A molecular score by quantitative PCR as a new prognostic tool at diagnosis for chronic lymphocytic leukemia patients. PLoS One. 5:e12780.
Ferrajoli A, et al. (2013) Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B-chronic lymphocytic leukemia. Blood. 122:1891–9.
Budoni M, et al. (2013) The immunosuppressive effect of mesenchymal stromal cells on B lymphocytes is mediated by membrane vesicles. Cell Transplant. 22:369–79.
Mahaweni NM, Kaijen-Lambers MEH, Dekkers J, Aerts JGJV, Hegmans JPJJ. (2013) Tumour-derived exosomes as antigen delivery carriers in dendritic cell-based immunotherapy for malignant mesothelioma. J. Extracell. Vesicles. 2:22492.
Rassenti LZ, et al. (2008) Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 112:1923–30.
Ito M, et al. (2014) MicroRNA-150 inhibits tumor invasion and metastasis by targeting the chemokine receptor CCR6 in advanced cutaneous T-cell lymphoma. Blood. 123:1499–511.
Wang M, Yang W, Li M, Li Y. (2014) Low expression of miR-150 in pediatric intestinal Burkitt lymphoma. Exp. Mol. Pathol. 96:261–6.
Yokobori T, et al. (2013) MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 104:48–54.
Sun Y, et al. (2013) Expression of miR-150 and miR-3940-5p is reduced in non-small cell lung carcinoma and correlates with clinicopathological features. Oncol. Rep. 29:704–12.
Ma Y, et al. (2012) miR-150 as a potential bio-marker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 61:1447–53.
Mraz M, et al. (2009) miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia. 23:1159–63.
Acknowledgments
This work was financed by the Télévie fund (Fonds de la Recherche Scientifique — Fonds National de la Recherche Scientifique [F.R.S.-FNRS]).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it.
The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this license, visit (https://doi.org/creativecommons.org/licenses/by-nc-nd/4.0/)
About this article
Cite this article
Stamatopoulos, B., Van Damme, M., Crompot, E. et al. Opposite Prognostic Significance of Cellular and Serum Circulating MicroRNA-150 in Patients with Chronic Lymphocytic Leukemia. Mol Med 21, 123–133 (2015). https://doi.org/10.2119/molmed.2014.00214
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2119/molmed.2014.00214